This article has been corrected. See "Erratum: Serological Evidence of Coxiella burnetii and SARS-CoV-2 Co-Infection: A Case Report" in Volume 42 on page 117.
Dear Editor,
The most common symptoms of coronavirus disease 2019 (COVID-19) are respiratory symptoms that are not easily distinguishable from those of other acute respiratory infections [1]. As bacterial/fungal co-infections are reported in 8% of COVID-19 patients, diagnosing them is critical for appropriate treatment [2]. Pathogens that cause co-infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) include influenza A/B viruses, Mycoplasma pneumoniae, Acinetobacter baumannii, Candida albicans, and Legionella pneumophila [2]. Q fever is asymptomatic in approximately 60% cases, but a flu-like illness with high fever, myalgia, headache, and cough, which lasts for one to three weeks, may occur in acute infection and then resolve spontaneously [3]. We report the case of a 37-year-old man who was diagnosed as having Coxiella burnetii and SARSCoV-2 co-infection. To the best of our knowledge, this is the first such report in Korea. The study was approved by the Institutional Review Board of the Chungbuk National University Hospital, Cheongju, Korea (IRB number: 2020-03-025).
A 37-year-old man presented to the Chungbuk National University Hospital in May 2020 with fever, cough, and sputum development, which had started three days before hospital presentation. The patient was a farmer, with no epidemiologic link to COVID-19-confirmed cases. Physical examination revealed a temperature of 38.8°C, blood pressure of 126/64 mm Hg, pulse rate of 95/minute, and respiratory rate of 18/minute. A complete blood count revealed a hemoglobin level of 151 g/L, white blood cell count of 2.32 × 109 cells/L (absolute neutrophil count 1.37 × 109 cells/L, absolute lymphocyte count 0.64 × 109 cells/L), and platelet count of 116 × 109/L. Other blood tests revealed elevated levels of C-reactive protein (50.5 mg/L) and lactate dehydrogenase (12.07 µkat/L) and slightly elevated levels of D-dimer (12.05 nmol/L), AST (1.12 µkat/L), and ALT (0.83 µkat/L). The patient’s prothrombin time and activated partial-thromboplastin time tests were within the reference ranges (Table 1). Chest X-ray showed no active lung lesion. A real-time reverse transcription (real-time RT-PCR) (Allplex 2019-nCoV Assay, Seegen, Seoul, Korea) test for SARS CoV 2 was performed on admission day. The result was negative in a naso/oropharyngeal swab and positive in sputum. Multiplex RT-PCR results for M. pneumoniae, Chlamydia pneumoniae, L. pneumophila, Bordetella pertussis, Streptococcus pneumoniae, Haemophilus influenzae (Allplex PneumoBacter Assay, Seegen) and influenza A/B virus (Sofia fluorescence immunoassay, Quidel, San Diego, CA, USA) were all negative. Considering the patient’s occupation, serological tests for C. burnetii, Leptospira interrogans, and Orientia tsutsugamushi were performed on admission day and were all negative.
The next day, SARS-CoV-2 RT-PCR test results were negative. To rule out a false-positive result in the first RT-PCR test, we reextracted and re-tested the original samples. After re-extraction, the sputum tested negative. In addition, serological antibody tests for SARS-CoV-2 infection were performed to rule out a false-positive RT-PCR result. A sandwich ELISA targeting the SARS-CoV-2 receptor binding domain (RBD) of the spike protein (SP) was conducted the next day. Briefly, the SARS-CoV-2 RBD antigen was attached to a 96-well plate and diluted serum was applied. After washing to remove unbound substance, the detection antibody (horseradish peroxidase-conjugated anti-human IgG or IgM) was added. After washing away excess detection antibody, the optical density at 450 nm in each well was measured using a microplate reader. The anti-RBD IgG antibody test was positive on the day after admission (Fig. 1). Based on the positive SARS-CoV-2 RT-PCR result in sputum and positive sandwich ELISA result, the patient was diagnosed as having SARS-CoV-2 infection. Follow-up serological tests showed patient seroconversion indicating C. burnetii infection. Therefore, co-infection with C. burnetii and SARS-CoV-2 was confirmed in the follow-up period.
Although the severity of COVID-19 varies from mild to lifethreatening, bacterial or fungal co-infection in COVID-19 patients increases the risk of mortality [4]. Therefore, clinicians should consider the variable clinical severity of COVID-19 and the possibility of co-infection, which may cause the same symptoms as COVID-19 but can aggravate the patient’s condition and require additional laboratory testing for diagnosis.
Real-Time RT-PCR is a standard method for diagnosing SARS-CoV-2 infection, as it gives minimal false-positive results [5]. Considering that negative conversion of real-time RT-PCR test results takes more than two weeks for SARS-CoV-2 infection [6], the patient might have had SARS-CoV-2 infection in the past. On days 15 to 29 of COVID-19, the sensitivity of real-time RT-PCR is 70.7%, whereas that of ELISA is 100% [7]. In serological tests for SARS-CoV-2, various target proteins, such as RBD, nucleocapsid protein, and SP, can be used, and, when these tests are used in combination with molecular tests, the sensitivity and specificity of COVID-19 diagnosis are increased [7]. In our case, a false-positive real-time RT-PCR result could not be ruled out, but SARS-CoV-2 infection was assumed, considering the results of additional serological tests. We believe that the negative conversion of the real-time RT-PCR result was due to a low viral load or virus remnant. Anti-SARS-CoV-2 IgM is less sensitive than IgG [8], and a negative IgM result on days 3 and 4 is considered false. It is necessary to further evaluate the diagnostic performance of serological tests for COVID-19.
In Q fever, serological tests have been used to diagnose acute infection, and seroconversion from negative to positive occurs one to three weeks after symptom onset [3]. Although viral loads do not differ between asymptomatic and symptomatic COVID-19 patients, our patient’s symptoms at the time of hospital presentation are more likely to have been due to Q fever [9]. Other laboratory results were nonspecific, but lymphopenia was notable on admission. Lymphopenia is rarely observed in Q fever but is common in COVID-19, for which it is a prognostic indicator [10].
To the best of our knowledge, this is the first report on C. burnetii and SARS-CoV-2 co-infection, and serologic testing played an important role in the diagnosis. For accurate COVID-19 diagnosis, clinicians should consider a multidisciplinary approach and utilize accurate and rapid diagnostic tools.
ACKNOWLEDGEMENTS
We thank all clinicians and laboratory technologists in Chungbuk National University Hospital and BioNano Health Guard Research Center.
Notes
AUTHOR CONTRIBUTIONS
Park HS designed the study and wrote the manuscript; Bae PK carried out the experiment and analyzed the data; Son BR collected the data; Jeong HW provided clinical information and discussed the manuscript; Shin KS designed the study and edited the manuscript. All authors have read the approved the final manuscript.
REFERENCES
1. Zha L, Shen J, Tefsen B, Wang Y, Lu W, Xu Q. 2020; Clinical features and outcomes of adult COVID-19 patients co-infected with Mycoplasma pneumoniae. J Infect. 81:e12–5. DOI: 10.1016/j.jinf.2020.07.010. PMID: 32652163. PMCID: PMC7342079.
2. Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. 2020; Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 71:2459–68. DOI: 10.1093/cid/ciaa530. PMID: 32358954. PMCID: PMC7197596.


3. Gikas A, Kokkini S, Tsioutis C. 2010; Q fever: clinical manifestations and treatment. Expert Rev Anti Infect Ther. 8:529–39. DOI: 10.1586/eri.10.29. PMID: 20455682.


4. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. 2020; Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 395:1054–62. DOI: 10.1016/S0140-6736(20)30566-3.



5. Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. 2020; Guidelines for laboratory diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann Lab Med. 40:351–60. DOI: 10.3343/alm.2020.40.5.351. PMID: 32237288. PMCID: PMC7169629.

6. Carmo A, Pereira-Vaz J, Mota V, Mendes A, Morais C, da Silva AC, et al. 2020; Clearance and persistence of SARS-CoV-2 RNA in patients with COVID-19. J Med Virol. 92:2227–31. DOI: 10.1002/jmv.26103. PMID: 32484958. PMCID: PMC7301002.


7. Lou B, Li T, Zheng S, Su Y, Li Z, Liu W, et al. 2020; Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 56:2000763. DOI: 10.1183/13993003.00763-2020. PMID: 32430429. PMCID: PMC7401320.
8. Zhang Z, Hou Y, Li D, Li F. 2020; Diagnostic efficacy of anti‐SARS‐CoV‐2 IgG/IgM test for COVID‐19: a meta‐analysis. J Med Virol. 93:366–74. DOI: 10.1002/jmv.26211. PMID: 32568413. PMCID: PMC7361902.


9. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. 2020; SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 382:1177–9. DOI: 10.1056/NEJMc2001737. PMID: 32074444. PMCID: PMC7121626.



10. Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, et al. 2020; Hematological findings and complications of COVID-19. Am J Hematol. 95:834–47. DOI: 10.1002/ajh.25829. PMID: 32282949. PMCID: PMC7262337.

Fig. 1
Serological diagnosis of co-infection with Coxiella burnetii and SARS-CoV-2. Serological tests for SARS-CoV-2 IgM/IgG anti-RBD and C. burnetii were positive, indicating co-infection.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RBD, SARS-CoV-2 receptor binding domain.
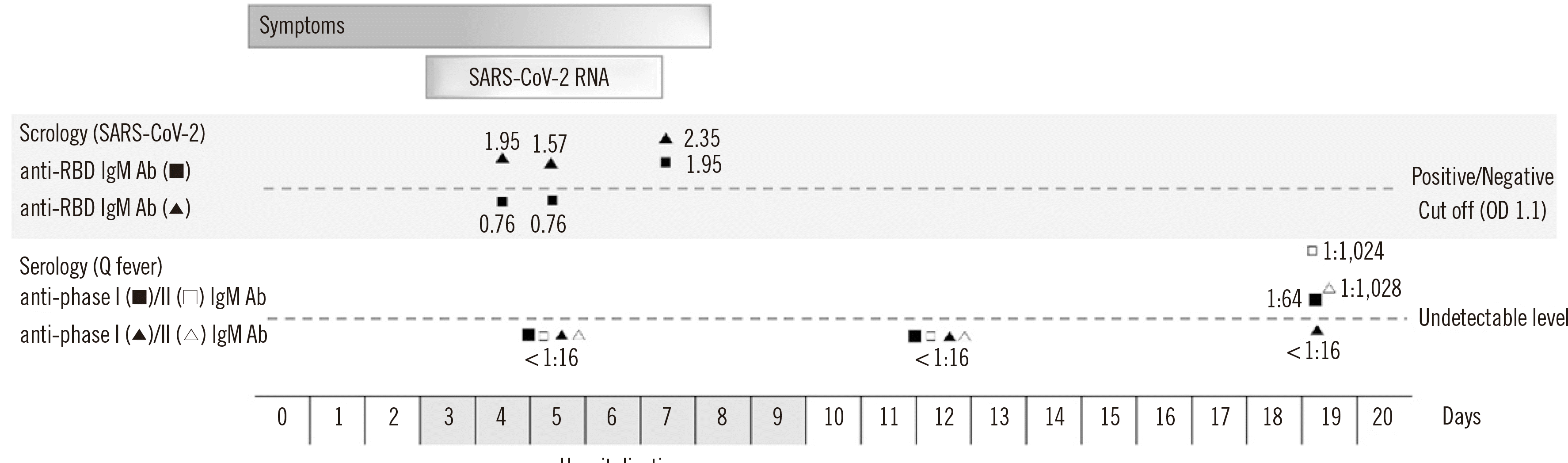
Table 1
Laboratory results after symptom onset
| Variable | Reference range | Day 3 | Day 4 | Day 5 | Day 6 | Day 19 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| NP&OP swabs | Sputum | NP&OP swabs | Sputum | |||||||||
|
|
|
|
|
|||||||||
| 1st/2nd /3rd* | Re-extraction | 1st/2nd /3rd* | Re-extraction | 1st | Re-sample | 1st | Re-sample | |||||
| Real time RT-PCR (Ct) | ||||||||||||
| E | < 40 | –/–/– | - | 32.79/33.03/33.88 | - | - | - | - | - | - | ||
| RdRp | < 40 | –/38.48/-– | - | 32.51/32.74/33.02 | - | - | - | - | - | - | ||
| N | < 40 | –/–/– | - | 34.94/35.54/36.30 | - | - | - | - | 39.69 | - | ||
| Hematologic parameter (unit) | ||||||||||||
| Hemoglobin (g/L) | 13–17 | 151 | 137 | 126 | 143 | |||||||
| White blood cells ( × 109/L) | 4–10 | 2.32 | 2.61 | 4.77 | 9.32 | |||||||
| Neutrophils ( × 109/L) | 1.37 | 1.06 | 1.99 | 2.70 | ||||||||
| Lymphocytes ( × 109/L) | 0.64 | 1.04 | 1.93 | 5.61 | ||||||||
| Platelets ( × 109/L) | 150–400 | 116 | 111 | 229 | 391 | |||||||
| PT (INR) | ≤ 1.2 | 0.99 | 1.02 | 1.07 | 0.96 | |||||||
| aPTT (sec) | 25.6–34.4 | 31.0 | 28.5 | 29.6 | 32.1 | |||||||
| CPK (µkat/L) | 0.97-5.81 | 14.83 | 7.83 | |||||||||
| LDH (µkat/L) | 4.39-7.51 | 12.09 | 10.47 | 10.32 | ||||||||
| D-dimer (nmol/L) | < 2.74 | 12.05 | ||||||||||
| CRP (mg/L) | ≤ 3.0 | 5.05 | 36.2 | 1.7 | ||||||||




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download