Botulism is a neuroparalytic disease caused by the seven immunologically distinct toxins (A to G) produced by
Clostridium botulinum, a gram-positive, anaerobic, spore-forming bacterium [
1,
2]. Botulinum toxin types A, B, E, and F cause botulism in humans [
2]. There are three main forms of botulism: foodborne botulism, infant botulism, and wound botulism [
3]. From 2002 to June 16 2019, eight cases of human botulism have been reported in Korea, and all were presumed to be foodborne botulism [
4–
6]. In the USA, more than 1,500 cases of infant botulism have been confirmed between 1976 and 2007 [
7]. By contrast, in Korea, the first documented case of infant botulism was reported in 2019, after botulism was designated as a notifiable infectious disease in 2002 [
8]. In addition, no
C. botulinum strains with two toxin gene clusters have been reported in Korea. This study aimed to characterize the
C. botulinum strain CB-27 isolated from a stool sample from the first case of infant botulism in Korea. We found that the strain possessed two different toxin gene clusters, showing genetic diversity compared with other previously reported
C. botulinum strains.
On June 7, 2019, a 4-month-old Korean baby with suspected infant botulism was admitted to Ajou University Hospital, Suwon, Korea, and on June 13, clinical samples, including serum and stool samples, were submitted to the Korea Centers for Disease Control and Prevention (KCDC) to identify
C. botulinum and its toxins [
8]. We conducted a mouse bioassay to identify botulinum toxins in the stool and serum samples [
9]. Based on neutralization of sample toxicity by monovalent antitoxin A (National Institute for Biological Standards and Control, Potters Bar, UK), only the stool sample was positive for botulinum neurotoxin (boNT) A.
A small amount of the stool sample was inoculated into cooked-meat medium (Difco, Franklin Lakes, USA) and cultured under anaerobic conditions at 37°C for three days. The culture was mixed 1:1 with 100% ethanol and incubated at 37°C for 1 hour. Serial culture dilutions were plated on egg yolk agar under anaerobic conditions to isolate
C. botulinum. Five lipase-positive colonies were isolated, and DNA was extracted from each strain using a genomic DNA isolation kit (Intron Biotechnology, Seongnam, Korea). We analyzed botulinum toxin genes by real-time PCR using previously reported primers [
10,
11]. As all strains were positive for both
boNT/A and
boNT/B genes, we selected the first isolated strain, named “CB-27,” and cultured it in trypticase peptone glucose yeast extract medium (KisanBio, Seoul, Korea) at 37°C for 24 hours. This study was exempted from the approval by the Institutional Review Board of the KCDC because clinical bacterial strains were used.
The culture filtrate of strain CB-27 was diluted 1:10,000 in phosphate-buffered saline containing 0.2% gelatin and then analyzed using a mouse bioassay. The mice were injected with 0.05 international units of botulinum antitoxins A and B (National Institute for Biological Standards and Control, Potters Bar, UK). Botulinum antitoxin A and a mixture of antitoxins A and B showed neutralizing ability against the CB-27 culture filtrate (
Table 1) in accordance with the mouse bioassay results obtained using stool samples.
Table 1
Mouse bioassay to identify C. botulinum neurotoxin type produced by CB-27 strain
|
Number of mice alive / Number of mice tested after injection with style="background-color:#e4eaf6;" |
|
|
Untreated filtrate |
Filtrate plus antitoxin A |
Filtrate plus antitoxin B |
Filtrate plus antitoxin A/B mixture |
|
0/2 |
2/2 |
0/2 |
2/2 |

To confirm the discrepancy between the mouse bioassay and real-time PCR analysis results, genomic DNA was extracted from the CB-27 strain and subjected to whole genome sequencing using a PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA;
https://www.pacb.com) and an Ion S5 (Thermo Fisher Scientific, Waltham, MA USA;
https://www.thermofisher.com) sequencer. The toxin gene cluster sequences of CB-27 were assembled de novo using the Hierarchical Genome Assembly Process version 3 (Pacific Biosciences). High-quality Ion S5 reads were used to correct potential sequencing errors in the PacBio long reads in Proovread version 2.14 (
https://github.com/BioInf-Wuerzburg/proovread) [
12]. The cluster sequence of CB-27 (67,538 bp) has been deposited at GenBank under accession number MT199282. Genes were annotated using rapid prokaryotic genome annotation (Prokka version 1.14.5,
https://github.com/tseemann/prokka) [
13]. Gene annotation showed that the cluster sequence of CB-27 harbored a
boNT/A gene and a silent
boNT/B gene (boNT/A(B)).
To phylogenetically characterize the toxin gene clusters of CB-27, we downloaded sequence data of 240 strains representing different toxin types and subtypes from Pathosystems Resource Integration Center (PATRIC) (
https://www.patricbrc.org) [
14]. All-against-all pairwise sequence comparisons were conducted using Basic Local Alignment Search Tool (
https://blast.ncbi.nlm.nih.gov/) to determine the closest relatives of CB-27. Multiple sequence alignments were conducted using MAFFT version 7.453 (
https://mafft.cbrc.jp/alignment/software/), with a maximum of 1,000 iterations. We constructed maximum-likelihood phylogenetic trees using RAxML-NG version 0.9.0 (
https://github.com/amkozlov/raxml-ng) with TVM+F+G4, the best nucleotide substitution model recommended by ModelFinder [
15,
16]. The
boNT/A nucleotide sequence of CB-27 was identical or highly similar to the sequences of previously reported subtype A1 strains, including CDC_69094 (100%) and ATCC 3502 (99.95%) (
Fig. 1A). The
boNT/B sequence of CB-27 showed 100% identity with that of CDC_69094 (
Fig. 1B), which contains a nucleotide substitution that can lead to premature termination of
boNT/B at amino acid position 128 [
17].
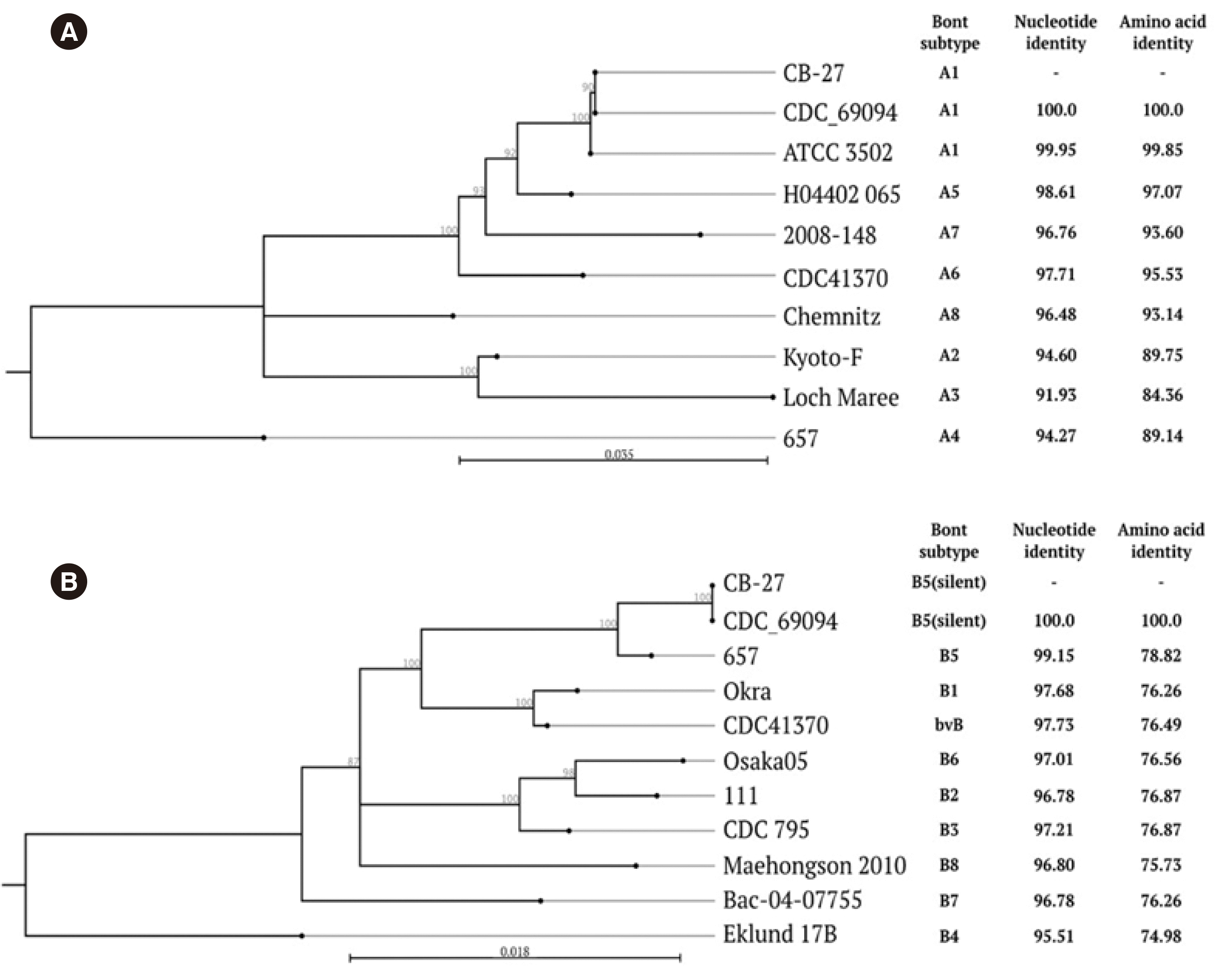 | Fig. 1Comparison of botulinum neurotoxin (boNT)/A and boNT/B nucleotide sequences. (A) The boNT/A sequences of strain CB-27 were compared with those of previously reported strains. (B) The boNT/B sequences of strain CB-27 were compared with those of previously reported strains. Nucleotide and amino acid identities of toxin genes between each strain and CB-27 are indicated on the right. The numbers on the tree indicate bootstrap values for branch points; only values >70 are shown. GenBank accession numbers are as follows: CDC_69094 (CP013246), ATCC 3,502 (AM412317), H04402 065 (EU679004), 2,008-148 (JQ954969), CDC41370 (FJ981696, FJ981697), Chemnitz (KM233166), Kyoto-F (X73423), Loch Maree (CP000963), 657 (CP001081), Okra (CP000940), Osaka05 (AB302852), 111 (AB084152), CDC 795 (EF028400), Maehongson 2010 (JQ964806), Bac-04-07755 (JQ354985), Eklund 17B (EF051570), and CB-27(MT199282). 
|
We examined genomic arrangements of the toxin gene clusters of CB-27 using the multiple genome alignment tool MAUVE version 2.4.0 (
http://darlinglab.org/mauve/mauve.html) [
18].
Fig. 2 shows a schematic diagram of the toxin A and B gene cluster structures of CB-27 and other representative strains (subtypes A1, A2, A3, and A(B)). The toxin gene cluster structures of CB-27 were identical to those of CDC_69094. Both CB-27 and CDC_69094 possess not only
ha70, ha17, ha33, botR, ntnh, and
boNT/A1 in the HA gene cluster but also
orfX3, orfX2, orfX1, botR, p47, ntnh, and
boNT/B5 (silent) in the OrfX gene cluster. However, the toxin gene cluster of CB-27 showed 99.53% nucleotide sequence identity with that of CDC_69094, showing genetic diversity between the toxin gene clusters.
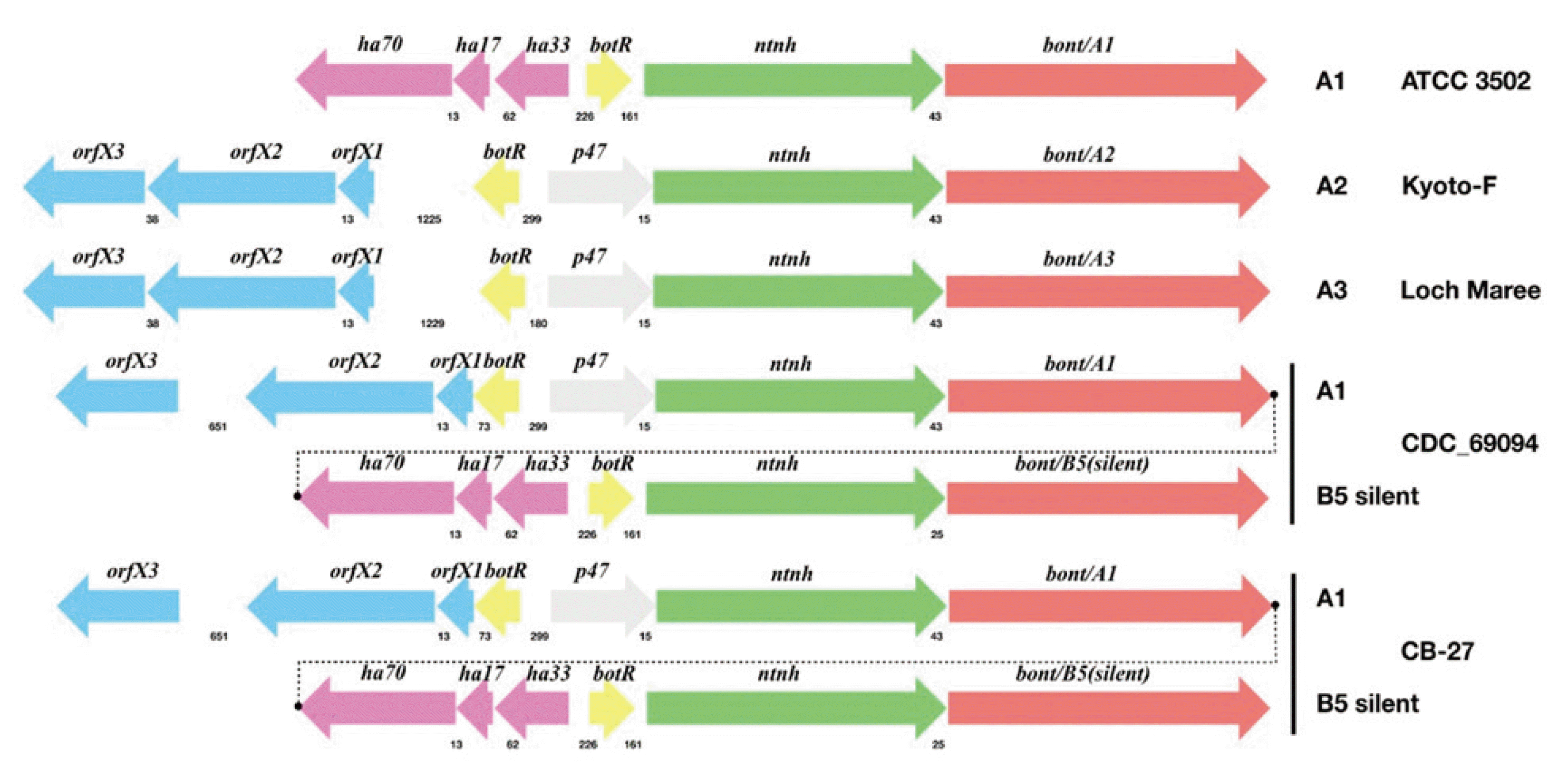 | Fig. 2The toxin gene cluster structures of CB-27 and other representative strains (subtypes A1, A2, A3, and A(B)). Gene locations and directions are indicated by arrows, with gene names presented above the arrows. The numbers between genes below the arrows indicate intergenic spacing. GenBank accession numbers are as follows: ATCC 3502 (AM412317), Kyoto-F (CP001581), Loch Maree (CP000963), CDC_69094 (CP013246), and CB-27(MT199282). 
|
In conclusion, we characterized the toxin gene clusters of C. botulinum strain CB-27 isolated from the first case of infant botulism in Korea in 2019. The CB-27 genome possesses boNT/A1 and unexpressed boNT/B5. Although the nucleotide sequences of the toxin gene cluster of CB-27 showed a 0.47% difference from that of CDC_69094, the sequence of the complete genome of CB-27 strain may show a larger difference. Further analysis using whole genome sequencing of CB-27 will be needed to confirm this notion.
Go to :







 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download