Dear Editor,
Targeted RNA-sequencing (RNA-seq) using next-generation sequencing (NGS) technology is a highly accurate method for selecting and sequencing specific transcripts of interest [
1]. We routinely applied a customized targeted RNA-seq system during the diagnostic phase of hematologic malignancies. Our system detected the first Korean case of
NUP98-NSD1 and a novel
SNRK-ETV6 fusion with therapy-related acute myeloid leukemia (t-AML) showing a dismal clinical course.
NUP98-NSD1 accounts for approximately 4% of pediatric AML cases and shows a poor prognosis [
2,
3]. It could be created by a cryptic t(5;11)(q35;p15.5) and exerts a leukemogenic function by binding near the
HOX locus and
MEIS1 to increase expression via histone modifications [
4]. The Institutional Review Board of Chonnam National University Hwasun Hospital (CNUHH), Hwasun, Korea (CNUHH-2020-091) approved this study and granted a waiver of consent due to its retrospective nature. This report highlights the role of high-throughput parallel targeted RNA-seq in enhancing the diagnostic yield of hematologic malignancies.
In April 2020, a 14-year-old girl visited the outpatient clinic of CNUHH 1.5 years and 1.9 years after a matched unrelated peripheral blood stem cell transplantation and initial diagnosis of AML, respectively, for a follow-up bone marrow (BM) examination At initial diagnosis, the Korean AML 2012 regimen (double-induction strategy with idarubicin or mitoxantrone plus cytarabine, followed by consolidation therapy with cytarabine and etoposide) was administered and complete remission was achieved 28 days after the second induction. The laboratory findings showed a leukocyte count of 3.1×109/L, absolute neutrophil count of 0.58×109/L, hemoglobin of 114 g/L, and platelet count of 37×109/L. BM aspirates revealed 28% leukemic blasts corresponding to French-American-British (FAB) type M2. The BM karyotype was 45,XX,add(3)(p25),del(5)(q?),-12,add(12)(p13)[8]//46,XY[12], and the multiplex reverse transcription (RT)-PCR (HemaVision kit; DNA Technology, Aarhus, Denmark) finding was negative.
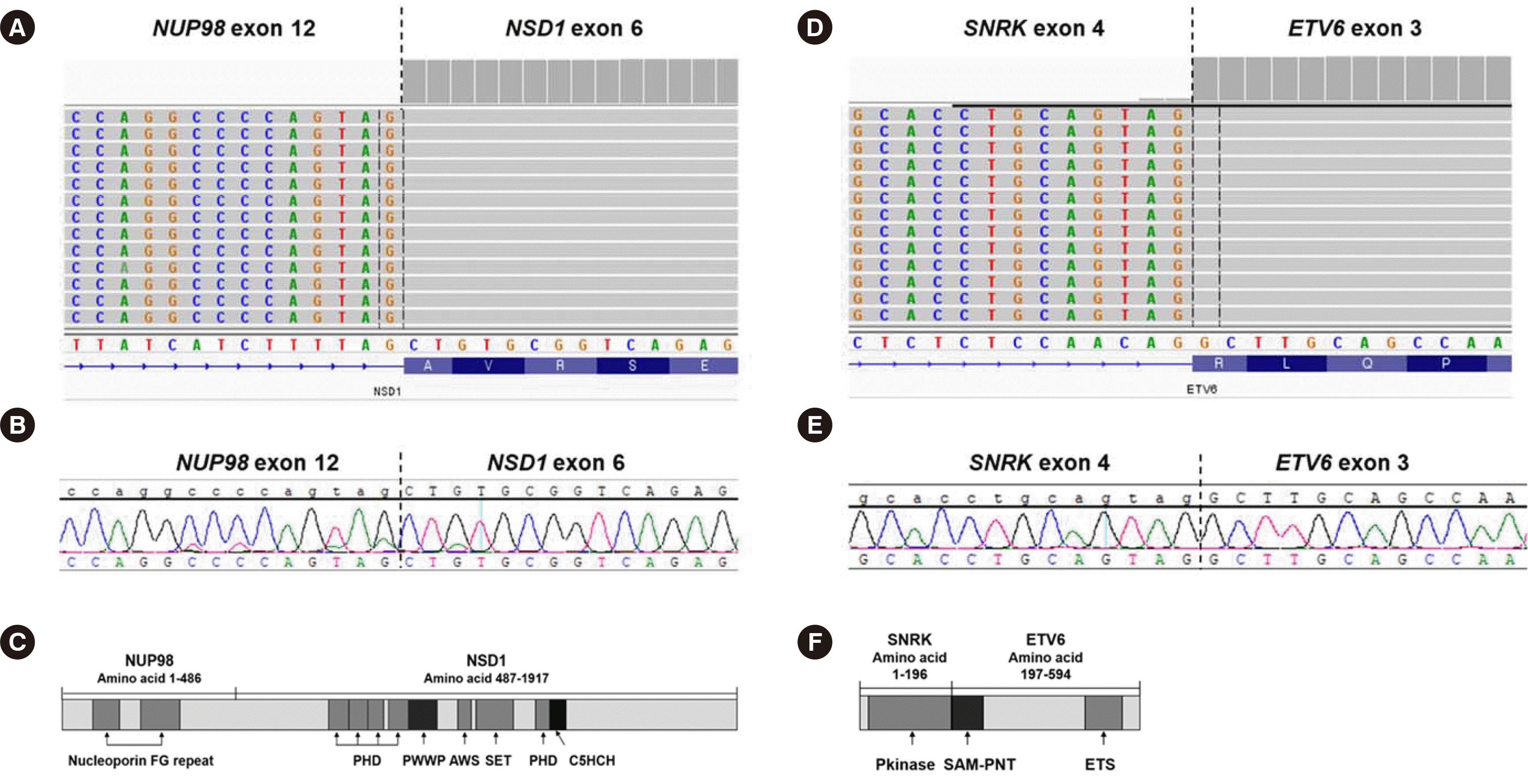
Targeted RNA-seq (HEMEaccuTest RNA; NGeneBio, Seoul, Korea) of the BM sample using STAR-Fusion (ver 1.8.1) and FusionCatcher (ver 1.20) revealed
NUP98-NSD1 and a novel
SNRK-ETV6 fusion, which were confirmed by direct sequencing (
Fig. 1). DESeq2 (ver 1.18.1) analysis showed that
WT1,
ERG, and
BAALC expression increased 7.1, 5.6, and 4.1-log
2-fold, respectively, compared with 14 normal controls (
Table 1). An additional tier II variant of
WT1, NM_024426.3:c.1142C>A (p.Ser381*), and three tier III variants were detected by FreeBayes (ver 1.3.1) [
5]. Further targeted DNA NGS (HEMEaccuTest DNA) confirmed the variants in targeted RNA-seq and additionally detected a tier II variant of
KRAS, NM_033360.4:c.38G>A (p.Gly13Asp), and five tier III variants. However, no significant variant of
FLT3, including
FLT3-ITD, was detected. Donor lymphocyte infusion (DLI) was conducted on day 7 after the diagnosis; however, the BM blasts increased to 88% on day 29. The combination of fludarabine, cytarabine, idarubicin, and granulocyte colony-stimulating factor chemotherapy was started on day 35 and the BM blasts decreased to <5% on day 71 with sustained thrombocytopenia; however, the condition repeatedly relapsed on day 134 and the patient expired on day 223.
NUP98-NSD1+ AML is characterized by frequent FAB-type M4/M5, a normal karyotype, and
HOXA/B upregulation [
2]. Further,
NUP98-NSD1 is mutually exclusive with other type II variants, but often co-occurs with type I variants such as
FLT3-ITD or
WT1 variants [
2,
3].
FLT3-ITD is the most common variant in
NUP98-NSD1+ AML (unlike our case), and its prognosis is dismal. Recent studies showed the promising therapeutic effects of dasatinib and navitoclax combination therapy and preemptive DLI based on minimal residual disease for
NUP98-NSD1+/
FLT3-ITD
+ AML [
6,
7]. Regarding the novel
SNRK-ETV6 fusion, the defect in ETV6 is pathogenic in hematologic malignancies caused by rearrangement or deletions [
8]. However, the partner
SNRK gene defect at 3p22.1 has rarely been studied in hematologic malignancies but reportedly impacts hematopoietic cell proliferation and differentiation [
9]. Further studies are needed to clarify the role of this novel fusion. This case also meets the criteria of t-AML, representing del(5q) with a complex karyotype and prior cytotoxic chemotherapy; both
NUP98- and
ETV6- rearrangements were reported in t-AMLs [
10]. Additionally, the patient has a variant in
TP53 (rs1042522), known to increase the risk of developing therapy-related myeloid neoplasms. Owing to the retrospective nature of this study, the
NUP98-NSD1 and
SNRK-ETV6 status at the initial diagnostic phase could not be ascertained.
Compared with previous studies using multiple diagnostic methods to characterize
NUP98-NSD1+ AML [
2], the advantage of the present case was the use of RNA-seq, representing a simplified diagnostic step for gene fusion, expression, and gene variant analyses. Additionally, this system might help uncover novel genetic characteristics in leukemias in future larger-scale studies.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download