Normal karyotype acute myeloid leukemia (NK-AML) is caused by a variety of genetic abnormalities that result in various clinical outcomes [
1-
3]. New molecular markers are required to develop a more accurate risk stratification system for evaluating and aiding the treatment of NK-AML.
Phospholipase C (PLC) isozymes are widely distributed in mammals and play essential roles in cell growth, signaling, and the development of pathological conditions, such as cancer [
4-
6]. Although associations between various cancers and PLC have been described [
7-1
7], there has been no study relating NK-AML with PLC-β2 expression. Therefore, we investigated the clinicopathological implications of PLC-β2 expressionin NK-AML patients.
Immunohistochemistry (IHC) was employed to assess PLC-β2 expression in formalin-fixed, paraffin-embedded (FFPE) bone marrow (BM) sections. PLC-β2 expression was assessed in 101 patients newly diagnosed as having NK-AML from November 2010 to September 2016 at Chonnam National University Hwasun Hospital, Hwasun, Korea. This study was approved by the Institutional Review Board of Chonnam National University Hwasun Hospital (IRB No. 2009-35), which waived the need for informed consent.
FFPE BM sections of 3 μm thickness were deparaffinized in xylene at 65°C for 10 minutes, rehydrated with ethanol, and rinsed with phosphate buffered saline. Thereafter, the slides were inserted to a Benchmark GX automatic stainer (Ventana Medical Systems, Oro Valley, AZ, USA). Rabbit polyclonal anti-human PLC-β2 antibody (clone# ab176012; Abcam, Cambridge, MA, USA) was applied at a 1:50 dilution. Monoclonal rabbit antihuman IgG1–4 antibody (clone# EPR4421, Abcam) at a 1:500 dilution was used as a negative isotype control. PLC-β2-immunostained slides were scored as follows: a brown granular cytoplasmic staining was considered positive, and positivity was scored on a scale of 0–8 as the sum of a diffuseness score (0–5) and an intensity score (0–3) (
Supplemental Data Table S1).
The final positivity score was determined as a median of scores by three independent pathology experts (
Supplemental Data Fig. S1). Overall survival (OS) and event-free survival (EFS) were analyzed using each positivity score as a cutoff and a median follow-up duration of 17.8 months. After determining the optimal cutoff value in the survival analysis, the IHC results were compared with those collected from the medical records for the following prognostic markers:
NPM1 variants and
FLT3-ITD,
BAALC, and
WT1. OS was estimated using the Kaplan–Meier method. Unadjusted hazard ratios (HRs) were calculated from a univariate Cox proportional hazards model.
P<0.05 was considered significant.
Median patient age at diagnosis was 57 years (23–83 years). The male:female ratio was 57:44. Considering a positivity score of 7.0 as a cutoff, the PLC-β2 overexpression group showed higher OS (72.6% vs. 26.5%;
P=0.016) and lower HR (0.453;
P=0.019) than the PLC-β2 low expression group. In EFS analysis, the PLC-β2 overexpression group showed no significant survival gain (50.6% vs. 43.0%,
P=0.465) and HR change (0.7357;
P=0.464) (
Table 1 and
Fig. 1).
Fig. 1
Clinicopathological implications of PLC-β2 protein expression in NK-AML patients. (A) Overall survival of NK-AML patients based on PLC-β2 expression status using Kaplan–Meier analysis. NK-AML patients with PLC-β2 overexpression (IHC score ≥7) presented significantly longer overall survival, indicating that PLC-β2 serves as a good prognostic marker. (B) Hazard ratio and overall survival of NK-AML patients according to various prognostic factors. PLC-β2 protein is a good prognostic indicator, whereas FLT3-ITD variants are poor prognostic markers. Each hazard ratio was specified as an unadjusted hazard ratio by univariate analysis.
Abbreviations: PCL, phospholipase C; NK-AML, normal karyotype acute myeloid leukemia; IHC, immunohistochemistry; NPM1, nucleophosmin 1 gene; FLT3-ITD, FLT3 internal tandem duplication gene; WT1, Wilm’s tumor suppressor gene 1; BAALC, brain and acute leukemia, cytoplasmic gene.
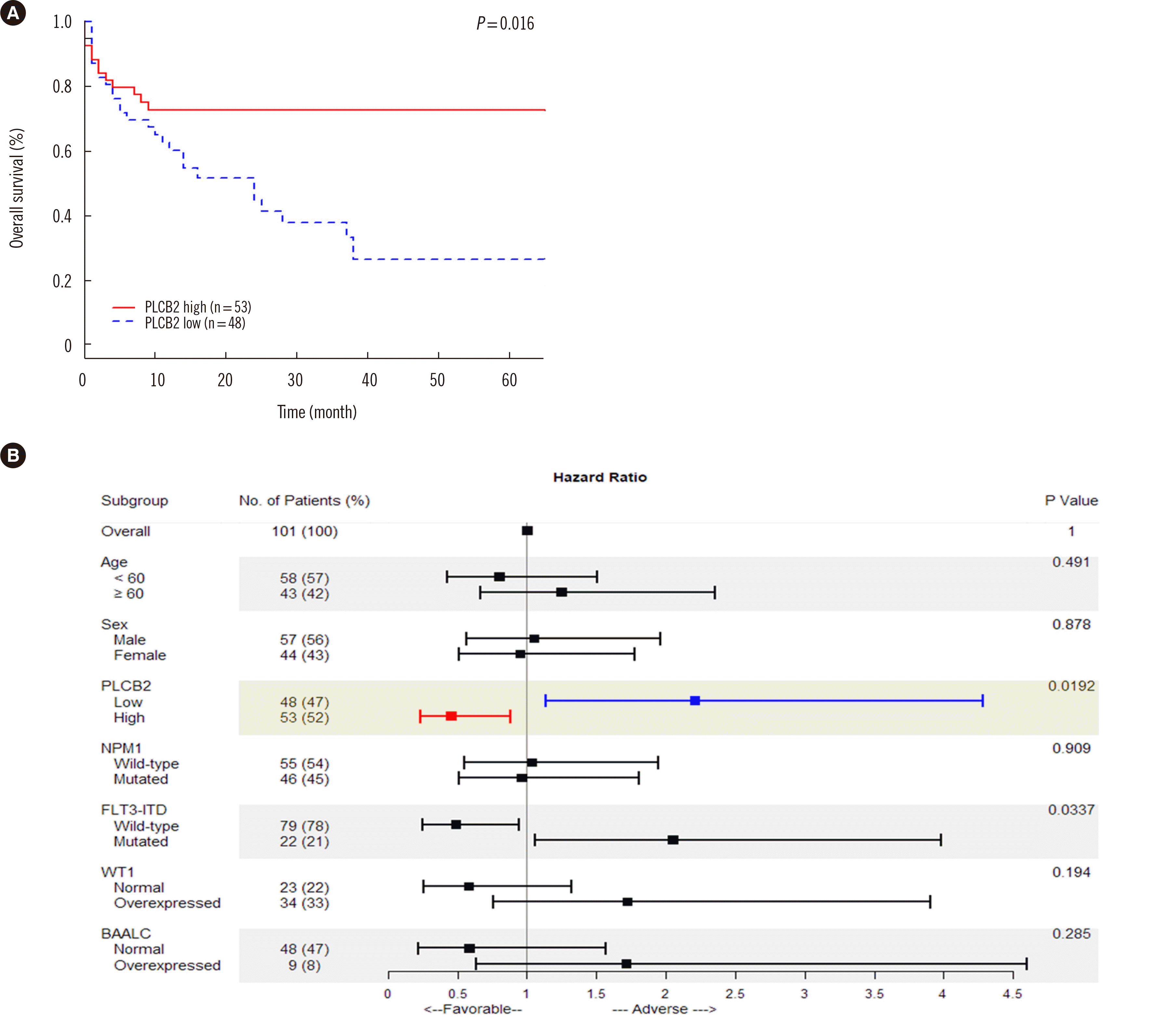

Table 1
Patient characteristics according to PLC-β2 protein expression
|
Patient characteristics |
Total (N = 101) |
PLC-β2 low expression (N = 48) |
PLC-β2 overexpression (N = 53) |
P
|
|
Age (yr) |
57 [49;66] |
56 [48;65] |
59 [50;68] |
0.496 |
|
Sex |
|
|
|
0.813 |
|
Male |
57 (56.4%) |
26 (54.2%) |
31 (58.5%) |
|
|
Female |
44 (43.6%) |
22 (45.8%) |
22 (41.5%) |
|
|
PLC-β2 IHC score |
7 [6;7] |
6 [4;6] |
7 [7;8] |
0.000 |
|
Overall survival (month) |
12.0 [3.0;26.0] |
22.8 [6.0;36.5] |
14.0 [2.0;26.0] |
0.016 |
|
Event-free survival (month) |
10.0 [3.0;21.0] |
10.0 [3.5;16.0] |
10.0 [2.0;24.0] |
0.943 |
|
PB WBC count (106/L) |
16,300 [3,800;59,290] |
12,150 [4,310;45,515] |
17,200 [3,330;84,200] |
0.357 |
|
Blast % of PB |
50 [10;80] |
50 [10;80] |
50 [10;80] |
0.913 |
|
Blast % of BM |
70 [50;80] |
70 [50;80] |
80 [60;90] |
0.180 |
|
Complete remission |
|
|
|
1.000 |
|
Achieved |
44 (44.9%) |
21 (44.7%) |
23 (45.1%) |
|
|
Failed |
54 (55.1%) |
26 (55.3%) |
28 (54.9%) |
|
|
Stem cell transplantation |
|
|
|
1.000 |
|
None |
63 (62.4%) |
30 (62.5%) |
33 (62.3%) |
|
|
Transplanted |
38 (37.6%) |
18 (37.5%) |
20 (37.7%) |
|

There were no significant differences in age, sex, white blood cell counts in peripheral blood (PB), blast percentage in PB and BM, complete remission rate, and enforcement of stem cell transplant between the PLC-β2 overexpression and low expression groups (
Table 1). Among the known prognostic markers, only
FLT3-ITD positivity was significantly associated with low OS (29.1% vs. negative group 52.7%;
P=0.032) and high HR (2.052;
P=0.034) (
Fig. 1) [
19].
NPM1-positive patients had a slightly, albeit not significantly, higher survival rate (
P=0.892) and lower HR 0.96 (
P=0.909). However, patients with
WT1 and
BAALC expression, which are poor prognostic factors, exhibited a slightly, albeit not significantly, lower survival rate (WT1
P=0.189,
BAALC P=0.280) and higher HR (WT1
P=0.194,
BAALC P=0.285) (
Fig. 1) [
1].
PLC-β2 expression decreased significantly in NK-AML patients, and the OS was significantly higher in the PLC-β2 overexpression group than in the PLC-β2 low-expression group (P=0.016). Given that the PLC-β2 pathway is involved in the regulation of platelet abundance, apoptosis, cell migration, and the cell cycle, these results suggest that PLC-β2 expression can be a potential prognostic factor.
AML cells produce anti-apoptotic factors in the cellular microenvironment that aid the survival of malignant cells, whereas the apoptosis of normal stem cells inhibits their survival [
18]. Inducing cellular apoptosis is one of the main functions of PLC-β2; it is presumed that apoptosis via PLC-β2 expression promotes an anti-apoptotic pathway-inducing microenvironment, leading to a poor prognosis in AML [
1].
In addition, PLC-β2 plays an essential role in cell migration mediated via intracellular and extracellular factors. PLC-β2 is an intracellular enzyme that mediates signal transduction from the C5a receptor to the C5 cut section (C5a) of the complement system, promoting granulocyte degranulation [
13]. A study on PLC-β2-knockout mice has revealed that PLC-β2-mediated protease degranulation and release are related to the normal migration of the intracellular enzyme from the BM to the PB [
13]. A decrease in PLC-β2 expression is thought to result from a diminished ability of the BM to release hematopoietic cells into the PB, resulting in a decreased cell survival rate.
The direct effects of PLC-β2 on biological functions (cell proliferation and apoptosis) may indicate that PLC-β2 expression can serve a useful indicator of normal neutrophil function in AML patients, and might, therefore, indicate a good prognosis in AML. Further, PLC release from blood cells during inflammation has been described in several pathological conditions; thus, PLC-β2 overexpression, which reflects a defense response in AML patients, may serve as an indicator of survival in these patients. Finally, PLC-β2 overexpression in NK-AML patients is presumed to be an essential indicator of the status of neutrophil cell cycle regulation. However, further study is needed to fully elucidate the functions and mechanisms of PLC-β2 in AML.
In conclusion, in addition to current prognostic indicators, PLC-β2 overexpression is an independent favorable prognostic indicator of NK-AML, as it is associated with high survival and low risk rates in NK-AML patients. Our results suggest that the PLC-β2 expressionassessed by IHC allows prognosis prediction in NK-AML.

