DISCUSSION
Considering the many fusion genes and breakpoint variants currently determined, more than 50 separate PCR reactions are required to screen an AL patient with a standard operation [
9]. We adopted a commercially accessible multiplex-RT-PCR system (DNA Technology) to identify 28 common fusion genes and over 80 splice variants based on previous research [
9]. Data from Denmark using this system, which was employed on specimens from 143 patients with a median age of 63 years (range, 0–85 yr; 132 adults, 11 children), revealed that CA was identified in only 15% (21/143) of the patients [
10]. In contrast, this study showed a higher CA frequency of 36% (228/637), using the same multiplex RT-PCR system: 35% (159/460) of AML patients, 38% (64/168) of ALL patients, and 56% (5/9) of MPAL patients. The rate of occurrence and spread of leukemic fusion genes in AL also differed from earlier European data [
10]. CA occurred in 65% (24/37) of childhood AML cases, whereas it only occurred in about 40% (24/60) of childhood AML cases based on the same multiplex RT-PCR system from Austria [
11]. The frequency of leukemic fusion genes in our ALL patients (38%, 64/168) was analogous to an Italian dataset utilizing the same multiplex RT-PCR system: 37% (28/75) and 39% (36/93) of childhood and adult ALL patients, respectively. In the Italian study, 39% of 170 ALL patients carried leukemic fusion genes: 39% and 40% of the childhood and adult ALL patients, respectively [
12].
The most common CA in AML patients was the t(15;17) abnormality (15%, 67/460). The occurrence rate was higher compared to data from other institutes of Korea (6.7%), Caucasian (6.5, 10%), Australian (12%), Japanese (11%), and Singapore-Chinese (11%), but it was analogous to the Chinese data (14.3%) [
5,
6,
13]. This discrepancy may be due to the sample size and likely to ethnic differences. The geographic diversity of CA in leukemia needs further study and a better understanding of the genetic and environmental factors associated with leukemia. The frequency and range of CA in ALL were similar between this study and previous studies [
6,
14]. A balanced translocation of the
BCR/ABL1 fusion gene in ALL patients was the most common abnormality (20%, 34/168) in this study. The occurrence of the
BCR/ABL1 fusion transcript was lower than in the Southwest Oncology Group data (26%) [
15], while it was higher than those in Indian (6%) and Taiwanese data (8%) [
16,
17]. The most common CA in childhood ALL patients was the
ETV6/RUNX1 fusion transcript, which was followed by
BCR/ABL1,
KMT2A gene rearrangements,
STIL/TAL, and
TCF3/PBX1. In contrast, the
BCR/ABL1 fusion transcript was the most common CA in adult ALL patients, which was followed by
TCF3/PBX1,
KMT2A/AFF1,
SET/NUP214, and
STIL/TAL1.
The discrepancy of 29% (159/553) between the multiplex RT-PCR system and conventional cytogenetics was mainly due to numerical and submicroscopic anomalies. The multiplex RT-PCR system used to screen 28 fusion transcripts identified 16 types in the group of 98 types of CA determined in 717 patients with leukemia. The concordance rate of 71% (394/553) between the multiplex RT-PCR system and conventional karyotyping was analogous to an earlier study (70%) [
1].
Choi
et al. [
1] reported 20 fusion transcripts containing KMT2A/EPS15 (
MLL1/AF-1p),
ETV6/ABL1 (
TEL/ABL),
RUNX1/MDS1 (
AML1/MDS1),
ZBTB16/RARA (
PLZF/RARA), and
ETV6/MN1 (
TEL/MN1) using a multiplex RT-PCR system. However, 16 fusion transcripts containing
DEK/ NUP214 were determined using the multiplex RT-PCR system in this study, and the above-mentioned 5 fusion transcripts were not identified:
KMT2A/EPS15 (
MLL1/AF-1p),
ETV6/ABL1 (
TEL/ABL),
RUNX1/MDS1 (
AML1/MDS1),
ZBTB16/ RARA (
PLZF/RARA), and
ETV6/MN1 (
TEL/MN1).
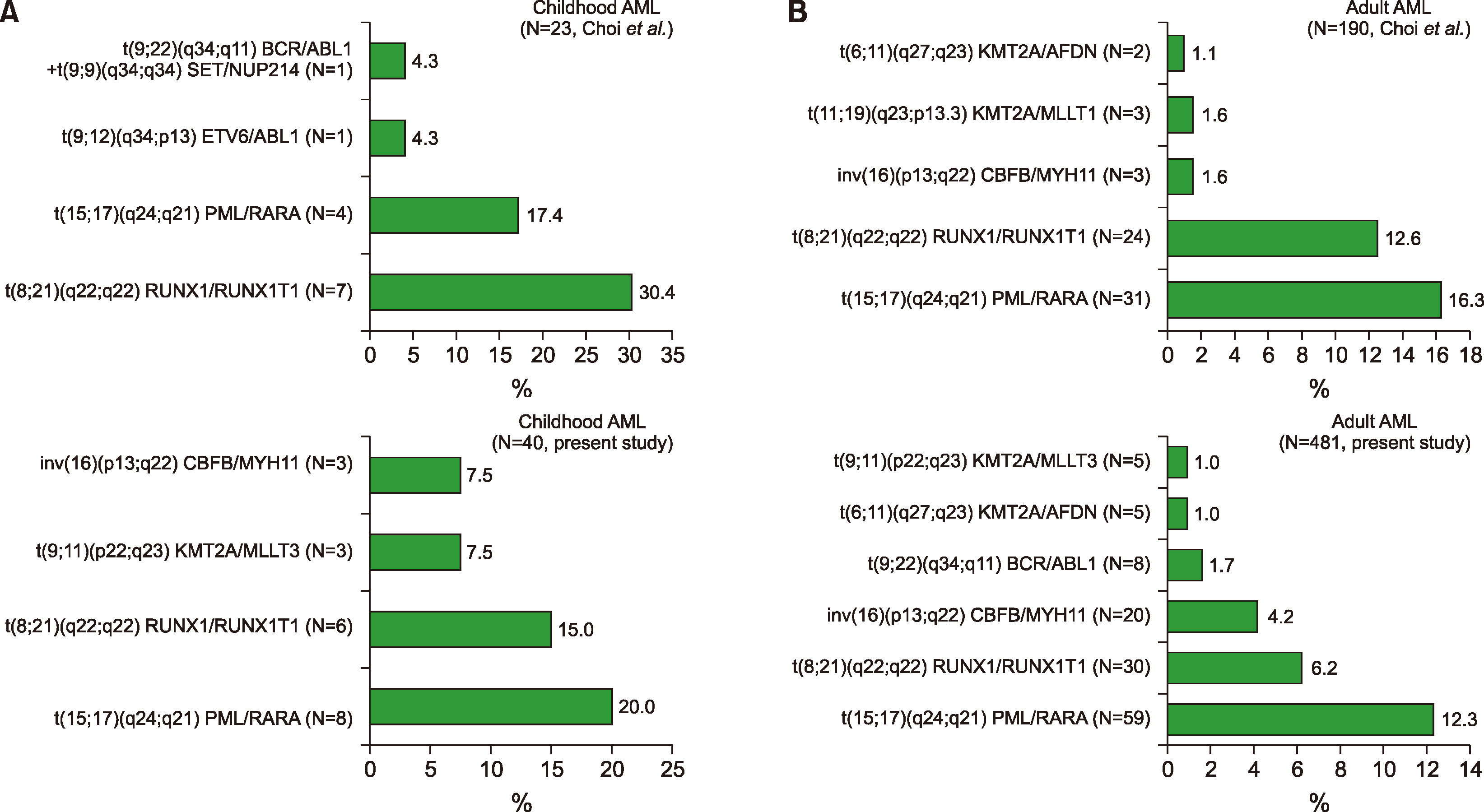
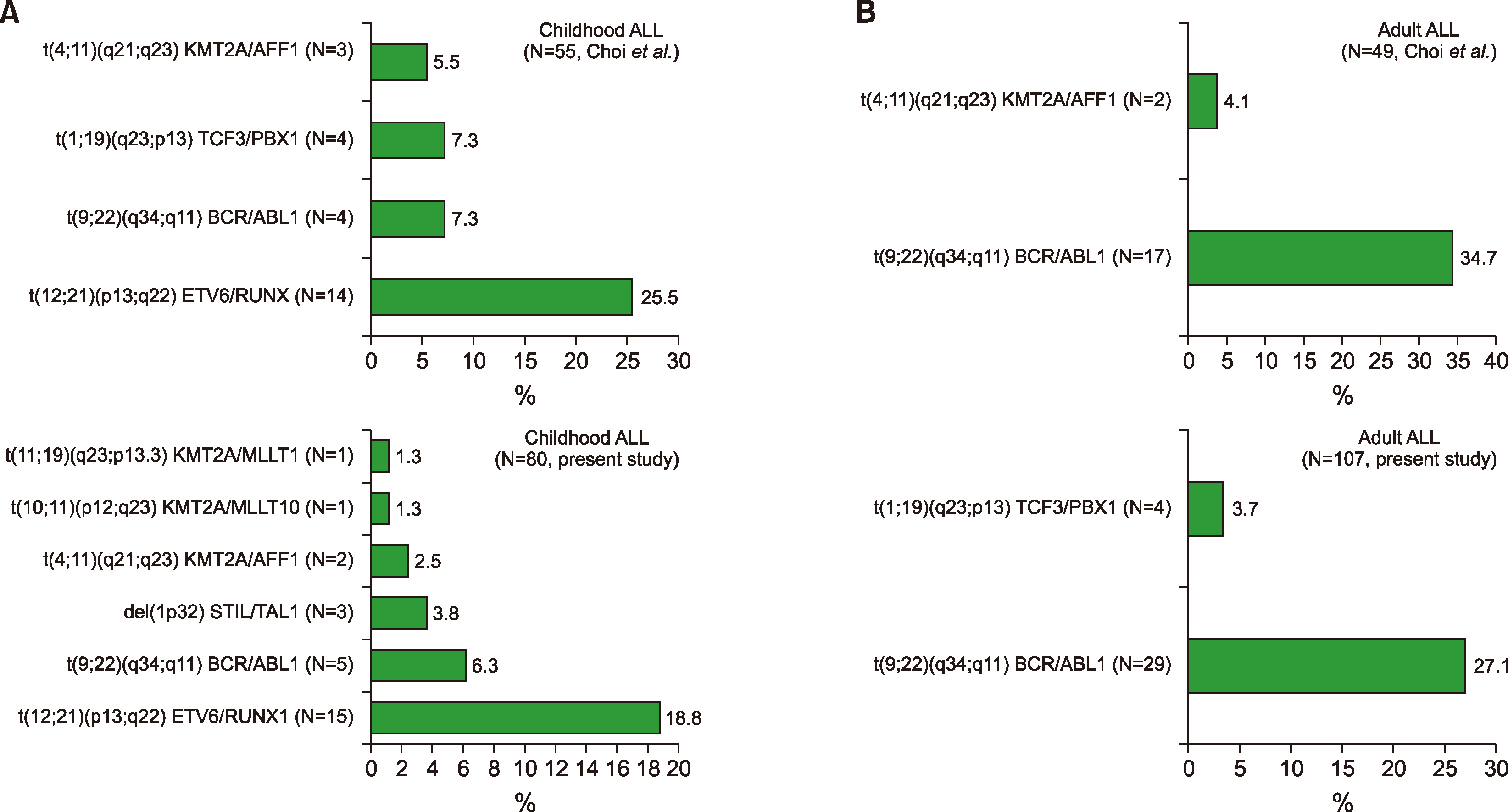
In addition, the range or spectra of leukemic fusion genes in AL differed from a prior report in Korea (
Fig. 1,
2) [
1]. In a prior study, Choi
et al. examined 325 leukemia patients during a 4-year period (2006–2009). Eighty-one children (median age 8 yr, range 0–18 yr) and 244 adults (median age 56 yr, range 19–86 yr) were included in their study. Of the 81 children, 35 (43%) were male and 46 (57%) were female; of the 244 adults, 123 (50%) were male. A total of 213 (66%) patients were diagnosed with
de novo (92%) or secondary (8%) AML (23 children, 190 adults); 104 (32%) patients had ALL (55 children, 49 adults), and 8 had mixed phenotype acute leukemia (MPAL). Accordingly, these findings were somewhat heterogeneous and different from those of the present study, as mentioned in the Patients’ section of the materials in this study.
In addition, the most prevalent CA in childhood AML patients was
PML/RARA, and was followed by
RUNX1/ RUNX1T1 in this study (
Supplementary Table 9), whereas the most prevalent CA was
RUNX1/RUNX1T1, which was followed by
PML/RARA in a previous report [
1]. In addition, taking into account MPAL patients, the most prevalent CA was
BCR/ABL1, followed by
KMT2A/MLLT3, in this study (
Supplementary Table 9, 10), while the most common CA was
BCR/ABL1, followed by
KMT2A/ELL in a previous report [
1].
Interestingly, there were cases with significant CA, such as i(17)(q10), t(3;3)(q21;q26.2), and t(8;14)(q24;q32), that were not identified using the commercial multiplex RT-PCR system (
Supplementary Table 8,
Supplementary Fig. 2). These are very meaningful and typical CA in hematological malignancies, such as AL. t(8;14)(q24;q32) is a typical CA in patients with ALL (Burkitt type), and the t(3;3)(q21;q26.2) is a newly adopted recurrent CA in AML patients, in accordance with the 2008 WHO classification [
1]. In addition, i(17)(q10) is repeatedly shown in the blast crisis of Philadelphia- positive CML patients. As a solitary CA, i(17q) has also been demonstrated in primary fibrosis, hypereosinophilic syndrome, and rare cases of myelodysplastic syndrome (MDS) and myeloproliferative neoplasm (MPN) that result in acute non-lymphocytic leukemia. In accordance with several studies, MDS/MPN, which contains the i(17q) as a single CA, is regarded as a distinctive entity with a frequency of 0.4–1.57% of MDS, or 1% of all myeloid cases, which features a male predominance, significant anemia, hyposegmented neutrophils, increased micromegakaryocytes, and an unfavorable prognosis [
18,
19]. Thus, it is worth considering CA as an additional screening panel of leukemic fusion genes for improving the molecular detection system.
In conclusion, this study disclosed the frequency and range of CA in Korean AL patients, which differed from those of previous published reports. This change might be due to environmental and lifestyle changes [
7,
8]. Our data were collected from a single center in Korea; thus, the findings may not be generalizable to other institutions. In addition, the limited number of patients might preclude definitive conclusions on the AL changes in Korea. Further studies to accumulate more evidence are needed to determine the change in spectra or range of AL. Nonetheless, this study might provide important information for improving the molecular detection system in Korean AL patients.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download