Dear Editor,
Jumping translocations (JTs) are rare but recurrent cytogenetic abnormalities in hematological malignancies and solid tumors [1]. Of the JTs reported in hematological malignancies, those involving chromosome 3q are the second most frequently observed donor segment [2-4]. However, to date, the impact of JTs involving 3q13.3 on the pathogenesis of leukemia has not been elucidated [4]. We describe the first RNA-sequencing (RNA-seq) data of a de novo acute myeloid leukemia (AML) patient with a JT involving 3q13.3 and trisomy 8 to investigate the structural variation and altered gene expression associated with JTs involving 3q13.3. The Institutional Review Board (IRB) of Samsung Medical Center, Seoul, Korea, approved this study (IRB No. SMC 2020-02-145-001). Informed consent for genetic studies were obtained from the patient before his death.
The patient was a 72-year-old man who was referred for a bone marrow (BM) study at Samsung Medical Center in April 2018 owing to the observation of 30% blasts on a peripheral blood smear. The BM study revealed 66% of blast cells with monocytoid differentiation. The patient was diagnosed as having AML, not otherwise specified, subtype acute monocytic leukemia. The patient was treated with decitabine for induction therapy due to his age; however, he died during the re-induction therapy owing to leukostasis and tumor lysis syndrome.
Conventional karyotyping was performed as previously described [5], and fluorescence in situ hybridization (FISH) analysis was performed on interphase nuclei using the LSI RPN1 SpectrumGreen/MECOM SpectrumOrange Probe (Vysis, Abbott Park, IL, USA), according to the manufacturer’s instructions. Copy number variation was analyzed using the CytoScan Dx Assay (Affymetrix Inc., Thermo Fisher Scientific, Santa Clara, CA, USA), according to the manufacturer’s instructions.
RNA was extracted from a BM aspirate using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). Paired-end libraries were prepared using the TruSeq RNA Sample Prep Kit (V2; Illumina, San Diego, CA, USA) and sequenced using a HiSeq 2500 instrument (Illumina). The sequence reads were mapped using STAR (version 2.4.0; https:// github.com/alexdobin/STAR) and quantified using RSEM (version 1.3.1; https://github.com/deweylab/RSEM). RSEM was also applied to generate the normalized gene expression in terms of transcripts per million values and to quantify the isoform-level expression of LSAMP. RNA expression was analyzed using Student t-test for each gene, and P < 0.05 was considered signifi cant; the fusion transcript was detected using ChimeraScan (version 0.4.5) and deFuse (version 0.6.2). The fusion transcript detected by RNA-seq was confirmed by Sanger sequencing.
Conventional cytogenetic analysis of the BM aspirate demonstrated three related cytogenetically aberrant clones (Fig. 1). The jumping donor chromosome bands ranging from 3q13.3 to qter translocated to three recipient chromosome regions: 12q24.3, 13p13, and 18p11.3. The G-banding karyotypic results were: 47,XY,+8,der(18)t(3;18)(q13.3;p11.3)[10]/47,XY,+8,der(12) t(3;12)(q13.3;q24.3)[4]/47,XY,+8,der(13)t(3;13)(q13.3;p13) [4]/46,XY[2]. FISH analysis using RPN1/MECOM, a dual color dual fusion probe, detected 24.0% of cells (48/200) with three MECOM signals on 3q26.2.
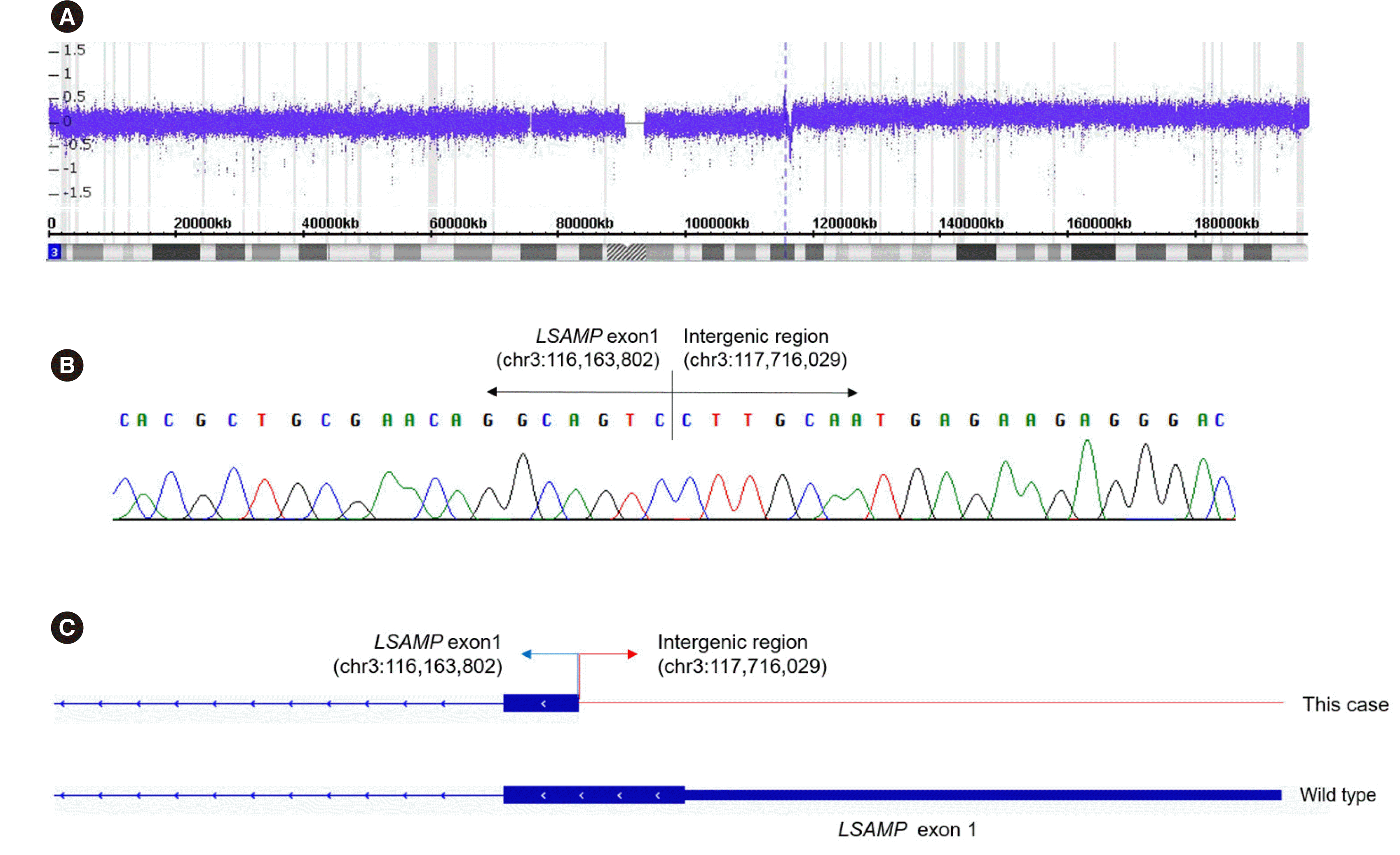
The chromosomal microarray showed partial trisomy of 3q13. 32-q29 (genomic position 117,967,114–197,851,986) and a single copy gain of 8p23.3-8q24.3 (genomic position 158,048– 146,295,771), which likely indicates trisomy 8. Moreover, a 385kb submicroscopic interstitial deletion at 3q13.31 (genomic position 116,234,782–116,620,270) was observed.
RNA-seq was performed to analyze the structural changes formed via the JT. A breakpoint resulting in a fusion between LSAMP exon 1 located on chromosome 3q13.31 and the intergenic region located at genomic position chr3:117,716,029 was observed (Fig. 2). The breakpoint was localized near the 3´ end of exon 1, spanning the 5´ untranslated region and the start codon of the LSAMP gene.
Gene expression was compared with the RNA expression data of 3q13.3 JT-negative AML patients. AML with a JT involving 3q13.3 showed significantly higher expression of LSAMP (P = 0.005). Quantification of the isoform-level gene expression of LSAMP revealed that isoform 1 transcripts were significantly increased (P = 0.003), whereas isoform 2 transcripts did not significantly differ. Of the genes located at the JT breakpoint, DL-GAP1, located on chromosome 18p11.31, showed overexpression. Overexpression of genes located on chromosome 3q (ACPP, NUDT16P, and EPHB3) or chromosome 8 (CHRNA6, ENPP2, MAPK15, and SDC2) was also observed.
JT-caused truncation or loss of the gene located near the donor chromosome breakpoint has been observed [6]. LSAMP is a putative tumor suppressor gene, previously reported to be inactivated by recurrent deletion and translocation in various cancer types [7-10]. Although LSAMP is mostly downregulated [7-10], Yen, et al. [9] reported that the correlation between LSAMP deletion and expression varied depending on the isoforms measured. In our study, LSAMP upregulation was detected, and its expression varied between its isoforms, suggesting alternative mRNA transcription or premature truncation. Our data showed the possibility that the rearrangement and aberrant LSAMP expression could be associated with the pathogenesis of AML with a JT involving 3q13.3.
Notes
AUTHOR CONTRIBUTIONS
Kim SH contributed to the conception and design of the study; Kim HY, Jang HY, Kim HJ, and Kim SH were involved in clinical evaluation; Cho EH and Kim HY interpreted the results; Yun JW performed the statistical analysis; Yoon J drafted the manuscript; and Kim SH supervised the study. All authors read and approved the final manuscript.
REFERENCES
1. Berger R, Bernard OA. 2007; Jumping translocations. Genes Chromosomes Cancer. 46:717–23. DOI: 10.1002/gcc.20456. PMID: 17444494.


2. Kjeldsen E. 2017; Characterization of an acquired jumping translocation involving 3q13.31-qter in a patient with de novo acute monocytic leukemia. Exp Mol Pathol. 103:14–25. DOI: 10.1016/j.yexmp.2017.06.002. PMID: 28625614.


3. Bernard M, Lemée F, Picard F, Ghandour C, Drenou B, Le Prise PY, et al. 2000; Jumping translocation in acute leukemia of myelomonocytic lineage: a case report and review of the literature. Leukemia. 14:119–22. DOI: 10.1038/sj.leu.2401637. PMID: 10637486.


4. Reis MD, Dubé ID, Pinkerton PH, Chen-Lai J, Robinson JB, Klock RJ, et al. 1991; "Jumping" translocations involving band 3q13.3 in a case of acute monocytic leukemia. Cancer Genet Cytogenet. 51:189–94. DOI: 10.1016/0165-4608(91)90131-D. PMID: 1993304.


5. Shin SY, Lee ST, Kim HJ, Cho EH, Kim JW, Park S, et al. 2016; Mutation profiling of 19 candidate genes in acute myeloid leukemia suggests significance of DNMT3A mutations. Oncotarget. 23:54825–37. DOI: 10.18632/oncotarget.10240. PMID: 27359055. PMCID: PMC5342384.
6. Hatakeyama S, Osawa M, Omine M, Ishikawa F. 1999; JTB: a novel membrane protein gene at 1q21 rearranged in a jumping translocation. Oncogene. 18:2085–90. DOI: 10.1038/sj.onc.1202510. PMID: 10321732.


7. Chen J, Lui WO, Vos MD, Clark GJ, Takahashi M, Schoumans J, et al. 2003; The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell. 4:405–13. DOI: 10.1016/S1535-6108(03)00269-1. PMID: 14667507.

8. Ntougkos E, Rush R, Scott D, Frankenberg T, Gabra H, Smyth JF, et al. 2005; The IgLON family in epithelial ovarian cancer: expression profiles and clinicopathologic correlates. Clin Cancer Res. 11:5764–8. DOI: 10.1158/1078-0432.CCR-04-2388. PMID: 16115914.


9. Yen CC, Chen WM, Chen TH, Chen WY, Chen PC, Chiou HJ, et al. 2009; Identification of chromosomal aberrations associated with disease progression and a novel 3q13.31 deletion involving LSAMP gene in osteosarcoma. Int J Oncol. 35:775–88. DOI: 10.3892/ijo_00000390. PMID: 19724913.


10. Petrovics G, Li H, Stümpel T, Tan SH, Young D, Katta S, et al. 2015; A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. EBioMedicine. 2:1957–64. DOI: 10.1016/j.ebiom.2015.10.028. PMID: 26844274. PMCID: PMC4703707.


FIGURES
Fig. 1
G-banding karyotype. Closed arrows indicate the JT breakpoints at each derivative chromosome. Karyotype showing (A) trisomy 8 and der(12)t(3;12)(q13.3;q24.3), (B) trisomy 8 and der(13)t(3;13)(q13.3;p13), and (C) trisomy 8 and der(18) t(3;18)(q13.3;p11.3). (D) Partial karyotype showing the JT in-volving 3q13.3.
Abbreviation: JT, jumping translocation.
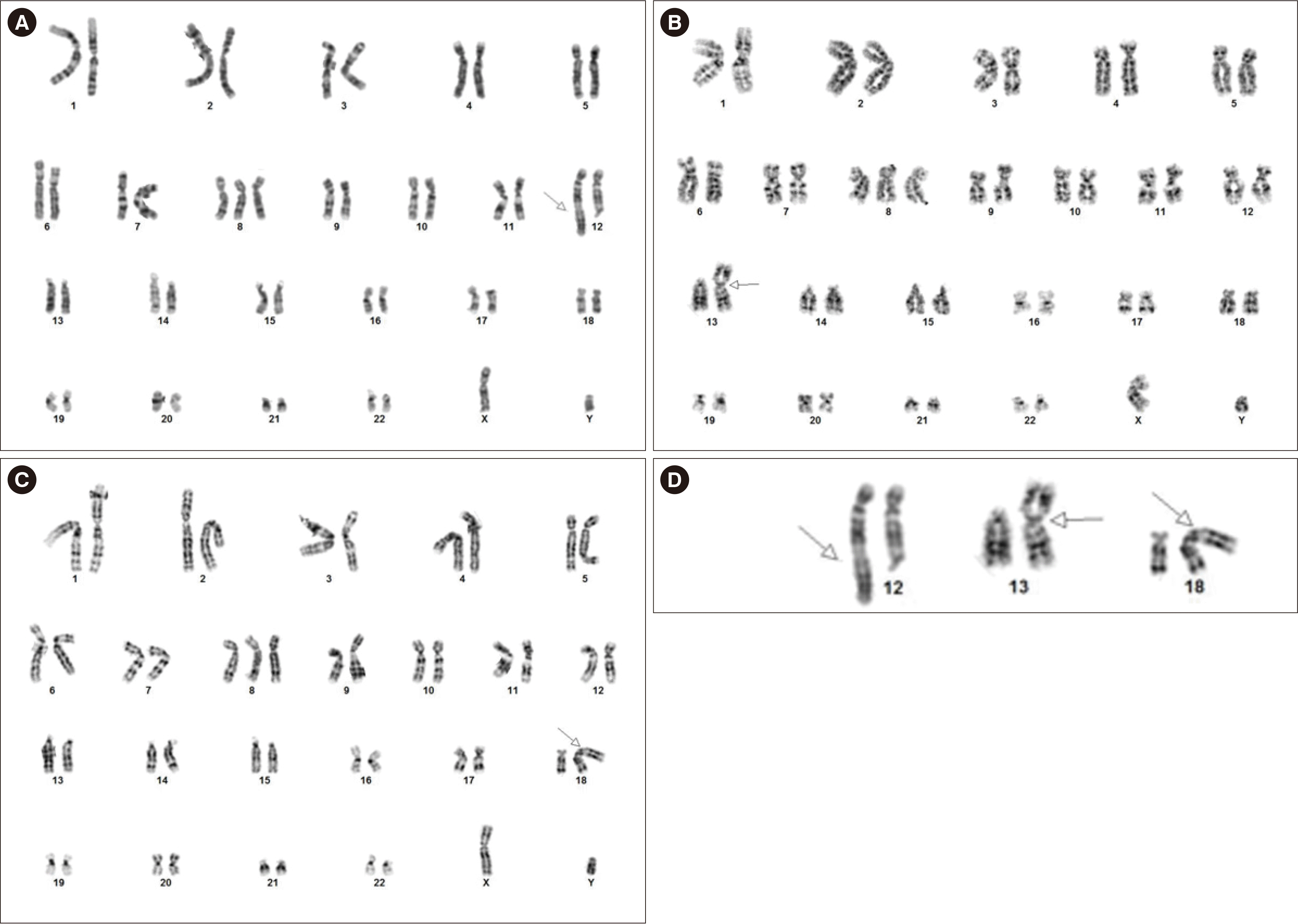
Fig. 2
The fusion transcript of the JT involving 3q13.3 identified by RNA-sequencing analysis and Sanger sequencing. (A) Chromosomal array demonstrating the breakpoints corresponding to an interstitial deletion of 3q13.31 and a gain in 3q13.32-q29. (B) Partial sequences surrounding LSAMP and the intergenic region fusion showing the junction of LSAMP exon 1 and the intergenic region (chr3:117,716,029). (C) Schematic diagrams of genomic breakpoints mapped with blue and red arrows indicating each breakpoint. The thick blue boxes represent the coding exon and 5´ untranslated region. The thin red line represents the DNA introns and intergenic regions.
Abbreviation: JT, jumping translocation.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download