Abstract
Background
Recent studies have successfully implemented next-generation sequencing (NGS) in HLA typing. We performed HLA NGS in a Korean population to estimate HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies up to an 8-digit resolution, which might be useful for an extended application of HLA results.
Methods
A total of 128 samples collected from healthy unrelated Korean adults, previously subjected to Sanger sequencing for 6-digit HLA analysis, were used. NGS was performed for HLA-A, -B, -C, and -DRB1 using the AllType NGS kit (One Lambda, West Hills, CA, USA), Ion Torrent S5 platform (Thermo Fisher Scientific, Waltham, MA, USA), and Type Steam Visual NGS analysis software (One Lambda).
Results
Eight HLA alleles showed frequencies of ≥10% in the Korean population, namely, A*24:02:01:01 (19.5%), A*33:03:01 (15.6%), A*02:01:01:01 (14.5%), A*11:01:01:01 (13.3%), B*15:01:01:01 (10.2%), C*01:02:01 (19.9%), C*03:04:01:02 (11.3%), and DRB1*09:01:02 (10.2%). Nine previous 6-digit HLA alleles were further identified as two or more 8-digit HLA alleles. Of these, eight alleles (A*24:02:01, B*35:01:01, B*40:01:02, B*55:02:01, B*58:01:01, C*03:02:02, C*07:02:01, and DRB1*07:01:01) were identified as two 8-digit HLA alleles, and one allele (B*51:01:01) was identified as three 8-digit HLA alleles. The most frequent four-loci haplotype was HLA-A*33:03:01-B*44:03:01:01-C*14:03-DRB1*13:02:01.
Go to : 
Human leukocyte antigen (HLA) is the most polymorphic gene among the known functional human genes [1]. Accurate identification of HLAs is important in solid organ and hematopoietic transplantations. Molecular HLA typing methods, including sequence-specific oligonucleotide probe, sequence-specific primers, and sequence-based typing methods have been widely used [2-4]. However, owing to the increasing number of HLA alleles, the problem of HLA typing ambiguity cannot be resolved, as these methods only analyze one or two exons where these variants are mainly found [5]. Recently, there have been several reports on successful implementation of next-generation sequencing (NGS) in HLA typing [6-8]. NGS has been reported to reduce ambiguity largely arising from heterozygotes [9]. By sequencing both exons and introns, NGS can reduce HLA typing ambiguity arising from sequencing only the specified regions [10, 11].
For high-resolution 8-digit HLA typing, we used the One Lambda AllType NGS Amplification kit (One Lambda, West Hills, CA, USA) to analyze the NGS results of HLA-A, -B, -C, and -DRB1 in a Korean population and established updated allele and haplotype frequencies, which will be useful for more precise and extended applications of HLA types in clinical and research fields including disease-related HLA type analysis, drug-related adverse reaction analysis, immunologic interaction studies, and anthropological genetic studies. Additionally, we compared our NGS results with previous Sanger sequencing results to identify any discrepancies between the two methods.
Go to : 
This retrospective study was performed at Asan Medical Center, Seoul, Korea, using the archival samples collected in 2003 from 128 genetically non-related, non-familial, healthy Korean adult volunteers > 18 years old. Samples were obtained by collecting 10–20 mL of venous blood from each subject, and the concentration of the extracted DNA was adjusted to 40–100 ng/µL. These samples were previously analyzed for 6-digit HLA types using Sanger sequencing with Big Dye Terminator 3.1 (ABI Inc., Foster City, CA, USA) and an ABI 3730 DNA analyzer (ABI Inc.) [12]. Samples were stored in a -70°C deep freezer until our analysis. The Institutional Review Board (IRB) at Asan Medical Center approved this study (IRB No. S2018-2423-0001) and waived the need for informed consents from subjects.
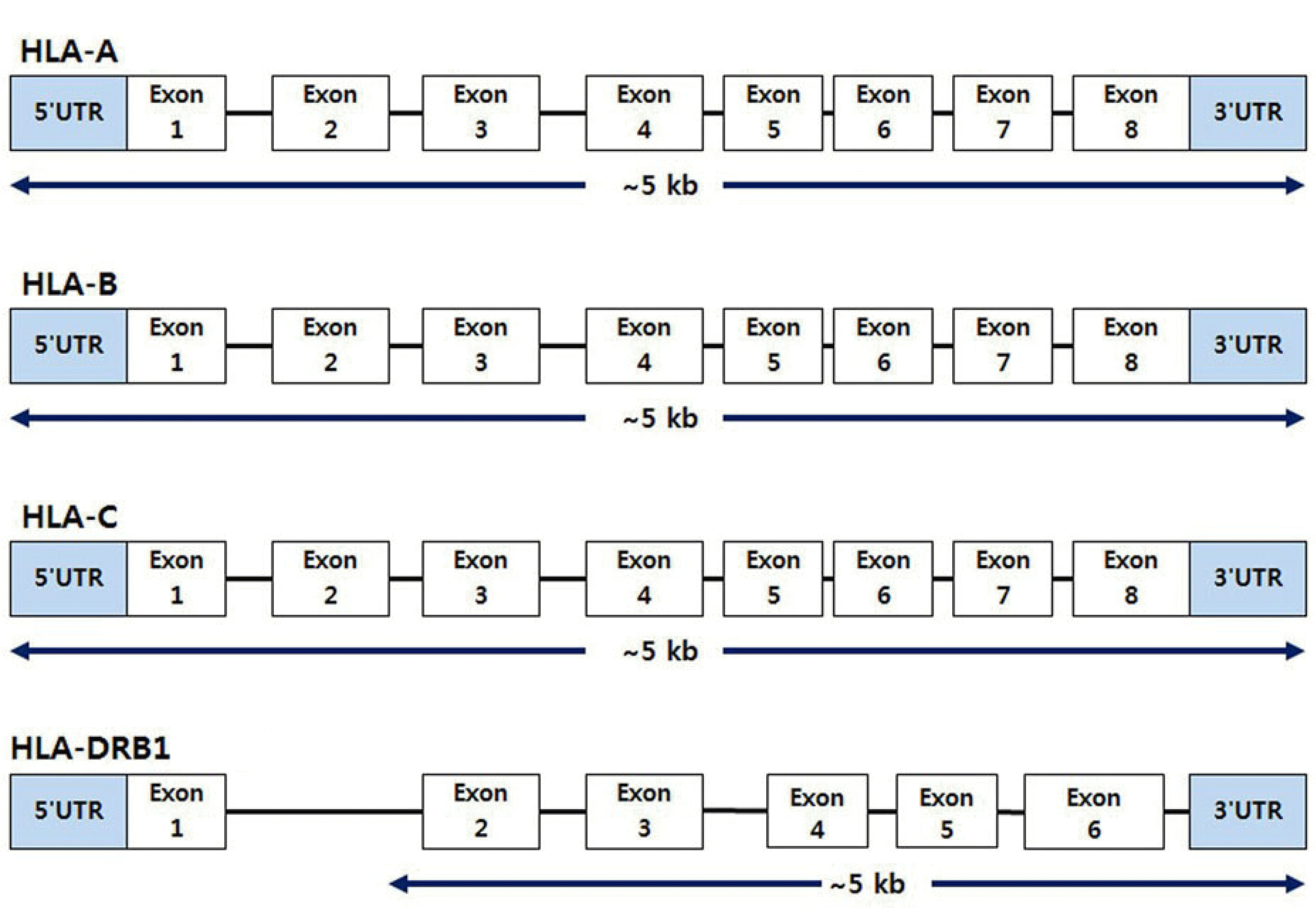
The AllType NGS 11-Loci Amplification Kit (One Lambda) was used to amplify target DNA regions. The reagents were mixed according to the number of samples and pipetted into a PCR plate containing DNA. PCR was then performed for 11 loci by multiplex tagging rather than per locus. The primers for PCR were provided by the manufacturer, and the PCR cycling condition was as follows: 1 cycle at 94°C for 2 minutes, 22 cycles of 10 seconds at 98°C, 3 minutes at 69°C, 8 cycles of 98°C for 10 seconds, and 3 minutes at 60°C. The coverage for each HLA-A, -B, -C, and -DRB1 locus is shown in Fig. 1. DNA was purified by mixing Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA, USA) with the amplicons. The purified amplicons were quantified using a Qubit 3.0 fluorometer and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions and were diluted to ensure equal concentrations. The amplicons were fragmented using the Ion Shear Plus Reagent Kit (Thermo Fisher Scientific) and then ligated by mixing the Ion Xpress Barcode Adapters and Ion Plus Fragment Library Kit (Thermo Fisher Scientific) in each well. Next, size selection using Agencourt AMPure XP Beads (Beckman Coulter) was performed. The final products were obtained using PCR after adding the Ion Plus Fragment Library Kit to the amplicon plate. The primers for PCR were provided by the manufacturer, and the PCR cycling condition was as follows: 1 cycle at 96°C for 62 minutes, 8 cycles of 96°C for 16 seconds, followed by 15 seconds at 58°C and 1 minute at 70°C.
Using the NGS Calculator Excel file provided by One Lambda, DNA amount was calculated and pooled. Clonal amplification and sequencing were performed using Ion 520 & 530 ExT Kit– Chef and Ion 530 Chip Kit (Thermo Fisher Scientific). Reagents were accurately positioned according to the manufacturer’s instructions; Ion Chef (Thermo Fisher Scientific) required 7 hours and Ion S5 XL (Thermo Fisher Scientific) required 6.5 hours. The obtained bam file was analyzed using Type Stream Visual (TSV) software (One Lambda). Analysis parameter settings are shown in Table 1, and the results were analyzed using the international ImMunoGeneTics information system (IMGT/HLA database) version 3.27 (the international ImMunoGeneTics information system, Montpellier, France).
Allele frequency was calculated using the Maximum Likelihood Estimation method of the ALLELE procedure with SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). All loci were evaluated to determine whether they meet Hardy-Weinberg equilibrium using the same program. Haplotype frequency was calculated with the Expectation-Maximization Algorithm of the HAPLOTYPE procedure using the same program. Statistical significance level was P < 0.05 (two-sided).
The base position of samples showing discrepant results between NGS and previous Sanger sequencing HLA genotypes obtained by Jun, et al. [12] was confirmed using the BIOWITHUS SBT Analyzer (Biowithus Inc., Seoul, Korea). Next, we performed PCR using primer sets (Avita Plus Kit, Biowithus Inc., Seoul, Korea) harboring the necessary additional exon and Sanger sequencing according to previous methods to confirm the sequence difference in the base position of the additional exon [12].
Go to : 
NGS HLA genotyping revealed 24 types of HLA-A, 44 types of HLA-B, 25 types of HLA-C, and 28 types of HLA-DRB1 in our study group; allele frequencies are shown in Table 2. A few 8-digit alleles (2 HLA-A, 11 HLA-B, 4 HLA-C, and 2 HLA-DRB1), previously reported as 6-digit genotypes were observed for the first time.
HLA-A, -B, -C, and -DRB1 allele frequencies in the Korean population (N=128)
HLA-A: previously typed 6-digit HLA-A*24:02:01 was further typed as two 8-digit HLA types of HLA-A*24:02:01:01 and A*24: 02:01:02L.
HLA-B: four of the previously typed 6-digit HLA-B alleles were further typed as two 8-digit HLA types (B*35:01:01 to B*35:01 01:02, B*35:01:01:06; B*40:01:02 to B*40:01:02:01, B*40:01:02:04; B*55:02:01 to B*55:02:01:01, B*55:02:01:02; B*58:01:01 to B*58:01:01:01, B*58:01:01:03), and previously typed 6-digit HLA-B*51:01:01 was further typed as three 8-digit HLA types of HLA-B*51:01:01:01, B*51:01:01:03, and B*51:01: 01:05.
HLA-C: two of the previously typed 6-digit HLA-C alleles were further typed as two 8-digit HLA types (C*03:02:02 to C*03:02: 02:01, C*03:02:02:03; C*07:02:01 to C*07:02:01:01, C*07:02:01:03).
HLA-DRB1: previously typed 6-digit HLA-DRB1*07:01:01 was further typed as two 8-digit HLA types of HLA-DRB1*07:01:01:01 and DRB1*07:01:01:02.
HLA haplotypes with frequencies > 1% are shown in Tables 3 and 4. There were 25 HLA-A-B-C haplotypes with >1% frequency, and the following were the most frequent haplotypes: HLA-A*33: 03:01-B*44:03:01:01-C*14:03, HLA-A*11:01:01:01-B*15:01:01:01-C*04:01:01:01, and HLA-A*33:03:01-B*58:01:01:03-C*03:02:02:01. There were 20 HLA-A-B-C-DRB1 haplotypes with > 1% frequency, and the following were the most frequent haplotypes: HLA-A*33:03:01-B*44:03:01:01-C*14:03-DRB1* 13:02:01, HLA-A*11:01:01:01-B*15:01:01:01-C*04:01:01:01DRB1*04:06:01, and HLA-A*02:01:01:01-B*13:01:01-C*03:04:01:02-DRB1*12:02:01.
HLA-A-B-C haplotype frequencies in the Korean population (N=128)
HLA-A-B-C-DRB1 haplotype frequencies in the Korean population (N=128)
A comparative analysis of the NGS and Sanger sequencing results revealed three discrepant HLA alleles (Table 5). HLA-A and -B alleles showed 100% concordance between the two methods, while there were two discrepant alleles for HLA-C and one discrepant allele for HLA-DRB1. Following additional SBT exon analyses of these samples, the SBT results were all corrected to the NGS results, yielding 100% concordance.
HLA allele discrepancies between NGS and 6-digit-resolution Sanger sequencing methods
Go to : 
We used the One Lambda AllType 11-loci multiplex kit to perform HLA NGS and obtained 8-digit HLA typing results in the Korean population.
The AllType NGS Amplification kit uses multiplex individual tagging of 11 HLA loci. Instead of detecting an independent locus at the PCR stage, the kit uses independent index combinations to detect all 11 loci from each sample. Thus, AllType has the following advantages: during the PCR stage only one step is needed, it requires less DNA and fewer pipetting events than other commercial kits, amplicon pooling is not needed, and the library preparation step is 8 hrs shorter, including the PCR stage. Eight of the HLA alleles showed frequencies of ≥ 10% in the Korean population. In previous studies examining HLA allele frequencies in a Korean population, In, et al. [14] reported five HLA alleles with frequencies of ≥10% [A*24:02 (22.7%), A*02:01 (17.6%), A*33:03 (16.1%), C*01:02 (17.8%), C*03:03 (11.8%)] by SBT, while Chung, et al. [15] reported seven HLA alleles [A* 24:02 (22.9%), A*02:01 (16.5%), A*33:03 (15.4%), B*51:01 (10.2%), C*01:02 (17.4%), C*03:03 (10.9%), DRB1*09:01 (10.4%)] by high-resolution DNA typing. Our results were mostly consistent with previous results in 4-digit resolution comparisons [14, 15]. Interestingly, B*51:01 (10.2%), reported by Chung, et al. [15], was divided into three 8-digit alleles, and C*03:03:01:01 was nearly 10% (9.4%) in our study.
A comparison of some of the most frequent HLA alleles in Koreans with those in individuals from other countries revealed high frequencies of HLA-A*24:02:01:01 (19.5%) in the Japanese (37.9%) and Hong Kong Chinese (14.7%) populations, whereas the frequencies were lower in US Caucasian (7.5%), British Caucasian (6.9%), and Saudi (8.5%) populations [16-20]. HLA-B*15:01:01:01 (10.2%) was not as frequent in other populations (Hong Kong Chinese 2.9%, US Caucasian 6.0%, British Caucasian 5.9%, and Saudi 0.3%) except in the Japanese population (8.7%). HLA-C*01:02:01 (19.9%) was also observed in significantly high frequencies in the Japanese (14.8%) and Hong Kong Chinese (19.0%) populations, whereas the frequencies were lower in the other populations (US Caucasian 2.1%, British Caucasian 4.0%, Saudi 1.0%). HLA-DRB1*09:01:02 (10.2%) was also significantly high in the Japanese (12.4%) and Hong Kong Chinese (15.9%) populations, but was low in the other populations (US Caucasian 1.2%, British Caucasian 1.6%, and Saudi 0.3%).
Numerous intron variants have been predicted to exist; however, identifying them was not regarded as cost-effective relative to their clinical importance [21]. However, HLA typing solely of exons can provide incomplete information for two reasons. First, regulatory elements in promoter genes or introns need to be identified to determine the difference in HLA expression, as this is related to disease phenotype [22, 23]. Several studies have reported that promoter or intron identification resolved the problems of null to low expression of certain HLA alleles [24-27]. Second, without complete gene sequencing, it would be difficult to identify disease causes and drug interactions arising from the high linkage disequilibrium in the HLA region. Thus, the clinical use of NGS for HLA typing is expected to increase in the era of precision medicine.
A limitation of this study is that we were unable to include 8-digit HLA allele frequencies of the DQ or DP loci. We observed numerous ambiguities at the 8-digit resolution in these loci, which could not be resolved by this study alone. As the IMGT database is constantly being updated, further analysis would be needed to report 8-digit DQ and DP loci without ambiguities. This is the first study on high-resolution 8-digit HLA typing using the One Lambda AllType NGS HLA Typing kit in the Korean population. We identified updated frequencies of HLA alleles and haplotypes by analyzing not only the exons but also the whole locus, including introns, 3´ untranslated regions (UTR), and 5´UTRs of HLAA, -B, -C, and by resolving ambiguities for HLA-DRB1. Our data can be used as additional information in identifying cases where the same 4-digit or 6-digit HLA types show different characteristics, especially in studies involving disease-related HLA type analysis, drug-related adverse reaction analysis, immunologic interaction studies, and anthropological genetic studies in the Korean population.
Go to : 
Notes
AUTHOR CONTRIBUTIONS
Choe W and Oh H-B planned the study; Choe W implemented the study; Chae J-D, Yang JJ, and Hwang S-H participated in the experiments and analysis; Choi S-E helped with statistical analysis; Choe W and Oh H-B wrote this manuscript.
RESEARCH FUNDING
This study was supported by EMBRI Grants 2018 SN0002 from the Eulji University, Korea.
Go to : 
REFERENCES
1. Borghans JAM, Keşmir C, et al. Flower D, Timmis J, editors. 2007. MHC diversity in individuals and populations. In silico immunology. Springer;Boston, MA: p. 177–95. DOI: 10.1007/978-0-387-39241-7_10.

2. Cao K, Chopek M, Fernández-Viña MA. 1999; High and intermediate resolution DNA typing systems for class I HLA-A, B, C genes by hybridization with sequence-specific oligonucleotide probes (SSOP). Rev Immunogenet. 1:177–208. PMID: 11253946.

3. Olerup O, Zetterquist H. 1992; HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 39:225–35. DOI: 10.1111/j.1399-0039.1992.tb01940.x. PMID: 1357775.


4. Saiki RK, Walsh PS, Levenson CH, Erlich HA. 1989; Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci U S A. 86:6230–4. DOI: 10.1073/pnas.86.16.6230. PMID: 2762325. PMCID: PMC297811.



5. Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M, Slavich L, et al. 2010; Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol. 71:1033–42. DOI: 10.1016/j.humimm.2010.06.016. PMID: 20603174.


6. Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, Trachtenberg EA, et al. 2009; High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 74:393–403. DOI: 10.1111/j.1399-0039.2009.01345.x. PMID: 19845894. PMCID: PMC4205125.



7. Erlich RL, Jia X, Anderson S, Banks E, Gao X, Carrington M, et al. 2011; Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 12:42. DOI: 10.1186/1471-2164-12-42. PMID: 21244689. PMCID: PMC3033818.



8. Holcomb CL, Höglund B, Anderson MW, Blake LA, Böhme I, Egholm M, et al. 2011; A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens. 77:206–17. DOI: 10.1111/j.1399-0039.2010.01606.x. PMID: 21299525. PMCID: PMC4205124.



9. Shiina T, Suzuki S, Ozaki Y, Taira H, Kikkawa E, Shigenari A, et al. 2012; Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens. 80:305–16. DOI: 10.1111/j.1399-0039.2012.01941.x. PMID: 22861646.


10. Hosomichi K, Jinam TA, Mitsunaga S, Nakaoka H, Inoue I. 2013; Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics. 14:355. DOI: 10.1186/1471-2164-14-355. PMID: 23714642. PMCID: PMC3671147.



11. Ehrenberg PK, Geretz A, Baldwin KM, Apps R, Polonis VR, Robb ML, et al. 2014; High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics. 15:864. DOI: 10.1186/1471-2164-15-864. PMID: 25283548. PMCID: PMC4196003.



12. Jun JH, Hwang K, Kim SK, Oh HB, Cho MC, Lee KJ. 2014; Estimation of the 6-digit level allele and haplotype frequencies of HLA-A, −B, and -C in Koreans using ambiguity-solving DNA typing. Tissue Antigens. 84:277–84. DOI: 10.1111/tan.12368. PMID: 24851935.


13. IPD-IMGT/HLA Statistics. https://www.ebi.ac.uk/ipd/imgt/hla/stats.html. updated on Nov 2019.
14. In JW, Roh EY, Oh S, Shin S, Park KU, Song EY. 2015; Allele and haplotype frequencies of Human Leukocyte Antigen-A, -B, -C, -DRB1, and -DQB1 from sequence-based DNA typing data in Koreans. Ann Lab Med. 35:429–35. DOI: 10.3343/alm.2015.35.4.429. PMID: 26131415. PMCID: PMC4446582.



15. Chung HY, Yoon JA, Han BY, Song EY, Park MH. 2010; Allelic and haplotypic diversity of HLA-A, -B, -C, and -DRB1 genes in Koreans defined by high-resolution DNA typing. Korean J Lab Med. 30:685–96. DOI: 10.3343/kjlm.2010.30.6.685. PMID: 21157157.


16. Saito S, Ota S, Yamada E, Inoko H, Ota M. 2000; Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 56:522–9. DOI: 10.1034/j.1399-0039.2000.560606.x. PMID: 11169242.


17. Kwok J, Guo M, Yang W, Lee CK, Ho J, Tang WH, et al. 2016; HLA-A, -B, -C, and -DRB1 genotyping and haplotype frequencies for a Hong Kong Chinese population of 7595 individuals. Hum Immunol. 77:1111–2. DOI: 10.1016/j.humimm.2016.10.005. PMID: 27769748.


18. Skibola CF, Akers NK, Conde L, Ladner M, Hawbecker SK, Cohen F, et al. 2012; Multi-locus HLA class I and II allele and haplotype associations with follicular lymphoma. Tissue Antigens. 79:279–86. DOI: 10.1111/j.1399-0039.2012.01845.x. PMID: 22296171. PMCID: PMC3293942.



19. Alfirevic A, Gonzalez-Galarza F, Bell C, Martinsson K, Platt V, Bretland G, et al. 2012; In silico analysis of HLA associations with drug-induced liver injury: use of a HLA-genotyped DNA archive from healthy volunteers. Genome Med. 4:51. DOI: 10.1186/gm350. PMID: 22732016. PMCID: PMC3698530.



20. Hajeer AH, Al Balwi MA, Aytül Uyar F, Alhaidan Y, Alabdulrahman A, Al Abdulkareem I, et al. 2013; HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in Saudis using next generation sequencing technique. Tissue Antigens. 82:252–8. DOI: 10.1111/tan.12200. PMID: 24461004.


21. Qu H, Fang X. 2013; A brief review on the Human Encyclopedia of DNA Elements (ENCODE) project. Genomics Proteomics Bioinformatics. 11:135–41. DOI: 10.1016/j.gpb.2013.05.001. PMID: 23722115. PMCID: PMC4357814.



22. Cocco E, Meloni A, Murru MR, Corongiu D, Tranquilli S, Fadda E, et al. 2012; Vitamin D responsive elements within the HLA-DRB1 promoter region in Sardinian multiple sclerosis associated alleles. PLoS One. 7:e41678. DOI: 10.1371/journal.pone.0041678. PMID: 22848563. PMCID: PMC3404969.

23. Thomas R, Apps R, Qi Y, Gao X, Male V, O'hUigin C, et al. 2009; HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 41:1290–4. DOI: 10.1038/ng.486. PMID: 19935663. PMCID: PMC2887091.



24. Dubois V, Tiercy JM, Labonne MP, Dormoy A, Gebuhrer L. 2004; A new HLA-B44 allele (B*44020102S) with a splicing mutation leading to a complete deletion of exon 5. Tissue Antigens. 63:173–80. DOI: 10.1111/j.1399-0039.2004.00134.x. PMID: 14705988.


25. Elsner HA, Bernard G, Eiz-Vesper B, de Matteis M, Bernard A, Blasczyk R. 2002; Non-expression of HLA-A*2901102 N is caused by a nucleotide exchange in the mRNA splicing site at the beginning of intron 4. Tissue Antigens. 59:139–41. DOI: 10.1034/j.1399-0039.2002.590212.x. PMID: 12028543.

26. Laforet M, Froelich N, Parissiadis A, Bausinger H, Pfeiffer B, Tongio MM. 1997; An intronic mutation responsible for a low level of expression of an HLA-A*24 allele. Tissue Antigens. 50:340–6. DOI: 10.1111/j.1399-0039.1997.tb02884.x. PMID: 9349616.

27. Tamouza R, El Kassar N, Schaeffer V, Carbonnelle E, Tatari Z, Marzais F, et al. 2000; A novel HLA-B*39 allele (HLA-B*3916) due to a rare mutation causing cryptic splice site activation. Hum Immunol. 61:467–73. DOI: 10.1016/S0198-8859(00)00108-7. PMID: 10773349.


Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download