서 론
재료 및 방법
1. 대상 환자
2. NF1 유전자 변이 검출 방법
1) mRNA 직접염기서열분석(Sanger sequencing)
2) Multiplex Ligation Dependent Probe Amplification (MLPA)
3. 통계
결 과
1. NF1 유전자 변이 빈도 및 분포
Table 1
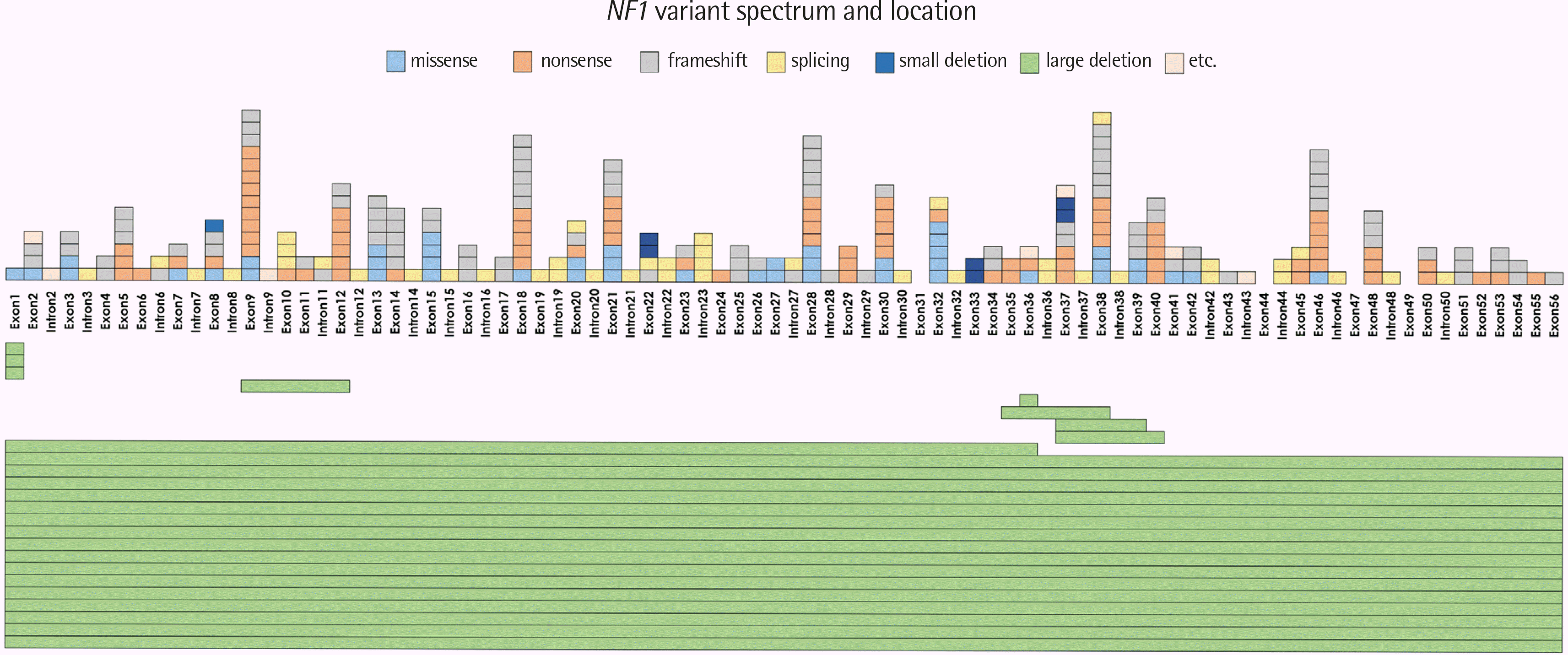 | Fig. 1Identified NF1 variant spectrum and location. At the midline of the figure, the involved exons and/or introns of the NF1 gene are listed. Each block upward the midline represents one variation. Different kinds of variations are shown in different colors as indicated in the category above the chart. Large deletions are indicated by the green rectangle at the bottom of the midline. Here, each variant is displayed in the vertical direction. The different horizontal length of each rectangle indicates the deleted scope of each individual variant. |
Table 2
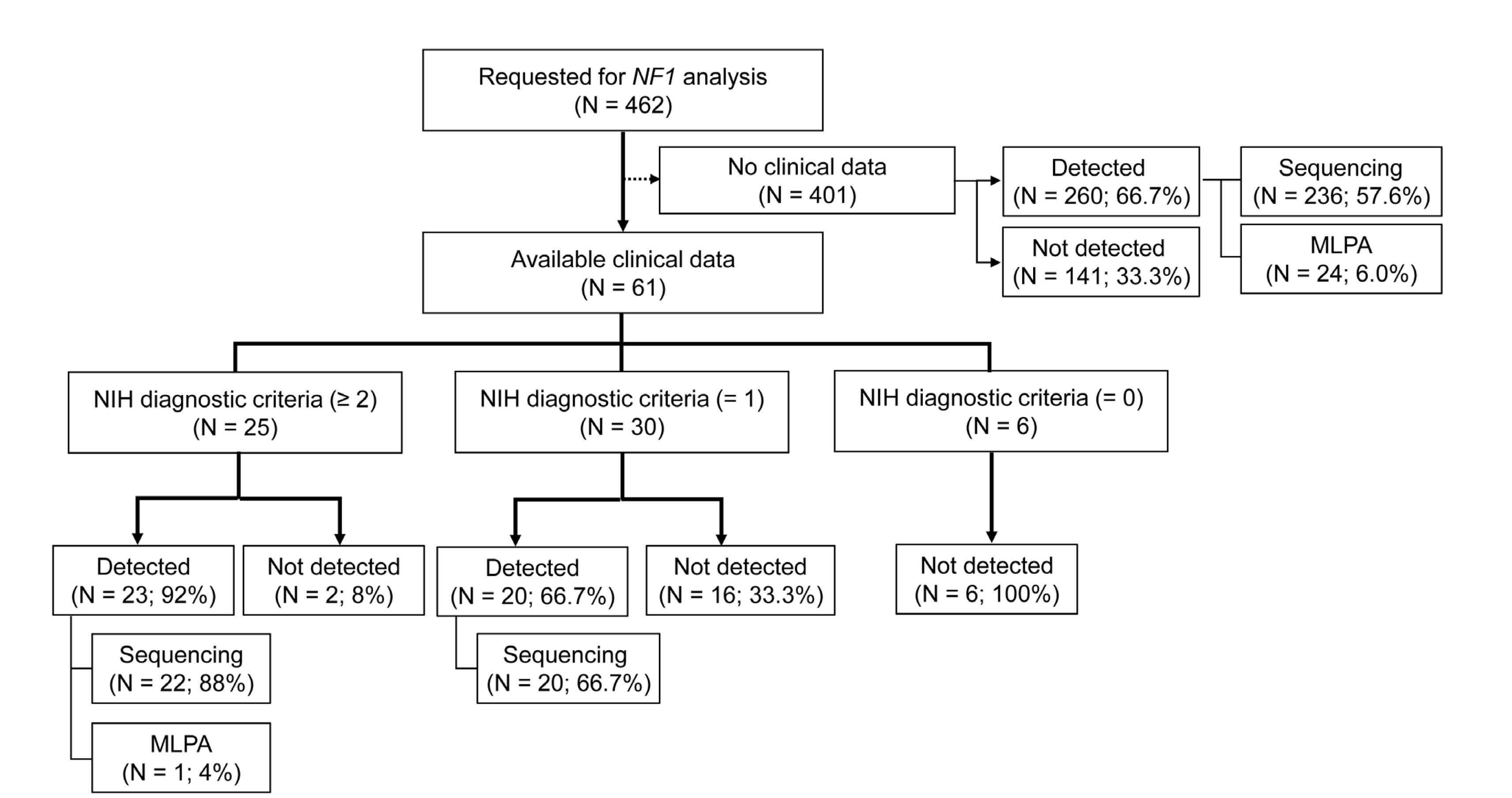 | Fig. 2Flow chart showing the NF1 gene analysis according to the clinical findings of enrolled patients. NIH, National Institutes of Health; Sequencing, Sanger sequencing of mRNA and gDNA; MLPA, multiplex ligation-dependent probe amplification. |
Table 3
| Mutation | Detected | Not detected | P | OR (95% CI) |
|---|---|---|---|---|
| Number of patients | 43 | 18 | ||
| Age, yr* | 13 (1-60) | 22 (4-74) | 0.014 | |
| Sex (N, %) | 0.678 | 0.792 (0.263-2.385) | ||
| Male | 24 (55.8%) | 9 (50%) | ||
| Female | 19 (44.2%) | 9 (50%) | ||
| Number of satisfying NIH criteria (1988) | ||||
| 2 or more | 23 (92.0%) | 2 (8%) | 0.002 | 9.200 (1.881-44.998) |
| One | 20 (66.7%) | 10 (33.3%) | < 0.001 | 4.583 (2.779-7.559) |
| None | 0 (0.0%) | 6 (100%) | ||
| Clinical presentation (N, %) | ||||
| 1st degree family history | 7 (16.3%) | 1 (5.6%) | 0.258 | 3.306 (0.376-29.045) |
| Café-au-lait spots | 39 (90.7%) | 9 (50.0%) | < 0.001 | 9.750 (2.446-38.869) |
| Neurofibroma | 18 (41.9%) | 1 (5.6%) | 0.005 | 12.240 (1.490-100.538) |
| Axillary/groin freckling | 4 (9.3%) | 1 (5.6%) | 0.627 | 1.744 (0.181-16.778) |
| Lisch nodule | 8 (18.6%) | 2 (11.1%) | 0.471 | 1.829 (0.348-9.603) |
| Optic glioma | 2 (4.7%) | 0 (0%) | 0.352 | 1.049 (0.982-1.120) |
| Skeletal dysplasia | 1 (2.3%) | 0 (0%) | 0.514 | 1.024 (0.978-1.072) |
| Brain hamartoma | 14 (32.6%) | 0 (0%) | 0.006 | 1.483 (1.205-1.825) |
| Seizure, mental retardation | 1 (2.3%) | 0 (0%) | 0.514 | 1.024 (0.978-1.072) |
| MPNST | 1 (2.3%) | 0 (0%) | 0.514 | 1.024 (0.978-1.072) |
| Other mass lesion | 0 (0%) | 6 (33.3%) | < 0.001 | 0.667 (0.481-0.924) |
고 찰
Table 4
| Country | Number of individuals | Methods | Detected numbers | Types of variants | Reference |
|---|---|---|---|---|---|
| Korea | 78 individuals from 60 families |
DNA sequencing mRNA sequencing MLPA FISH |
52 variants in 78 individuals |
Missense 23.1% Nonsense 34.6% Splicing 21.2% Indel 13.5% Large del 7.7% |
[13] |
| Spain | 474 unrelated individuals |
DNA sequencing mRNA sequencing mRNA SSCP/HD |
142 variants in 189 individuals |
Missense 8% Nonsense 9% Splicing 31% Frameshift 50% Others 2% |
[16] |
| China | 107 unrelated individuals |
DNA sequencing DHPLC microsatellite genotyping Q-PCR RT-PCR |
68 individuals |
Missense 16.2% Nonsense 29.4% Splice site 13.2% Frameshift 33.8% Large del 7.4% |
[17] |
| UK | 169 unrelated individuals |
DNA sequencing DHPLC MLPA FISH |
88 variants in 109 individuals |
Missense 15% Nonsense 27% Splicing 19% Frameshift 27% Large del 9% Others 3% |
[18] |
| France | 565 unrelated individuals |
DNA sequencing mRNA sequencing microsatellite genotyping MLPA |
546 individuals |
Missense 7.7% Nonsense 21.6% Splicing 27.1% Frameshift 30.0% Large del 8.4% Others 5.1% |
[19] |
| Japan | 86 unrelated individuals |
NGS DNA sequencing MLPA |
67 variants in 80 individuals |
Missense 16.3% Nonsense 37.5% Splicing 10% Frameshift 23.8% Large del 12.5% |
[20] |
| Netherlands | 1985 unrelated individuals |
DNA sequencing SSCP MLPA FISH |
1178 individuals |
Missense 9.8% Nonsense 29.4% Splicing 19.7% Frameshift 31.7% Others 4.1% |
[21] |
| China | 109 individuals, 100 families |
DNA sequencing mRNA sequencing MLPA |
89 variants in 100 families |
Missense 13.5% Nonsense 23.6% Splicing 16.9% Frameshift 29.2% Large del 12.4% Others 4.5% |
[22] |
| Italy | 79 unrelated individuals |
NGS MLPA |
73 variants in 79 individuals |
Missense 30% Nonsense 34% Splicing 7% Small del 24.7% Insertion 1.4% Large del 2.7% |
[23] |
Abbreviations: MLPA, multiplex ligation-dependent probe amplification; FISH, Fluorescence in situ hybridization; SSCP, single-strand conformation polymorphism; DHPLC, denaturing high performance liquid chromatography; RT-PCR, reverse transcriptase polymerase chain reaction; NGS, next-generation sequencing; Indel, insertion/deletion; del, deletion.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download