INTRODUCTION
The clinical diagnosis of leptospirosis is difficult to distinguish from that of other infectious diseases (e.g., malaria, dengue, rickettsial disease, Hantavirus, or acute viral disease), because the clinical manifestations and laboratory findings are nonspecific. Among the available diagnostic tools, a culture of the
Leptospira species and the microscopic agglutination test (MAT) are considered as the reference standard for the diagnosis of leptospirosis. However, both tests are not useful for early diagnosis. A culture of
Leptospira is difficult due to its slow growth rate and requirement for special media, and anti-
Leptospira antibodies can be detected 5–7 days after symptom onset.
12 Limmathurotsakul et al.
3 reported that the sensitivities of the culture, MAT, and culture plus MAT are low (10.5%, 49.8%, and 55.5%, respectively).
For a rapid and accurate diagnosis, the detection
Leptospira species DNA by the nested polymerase chain reaction (N-PCR) from human samples (blood, urine, or cerebrospinal fluid [CSF]) has been developed over the last two decades.
4 Herein, this case series describes the usefulness of N-PCR targeting the 16S rRNA gene in urine and CSF for the diagnosis of leptospirosis.
CASE DESCRIPTION
Case 1
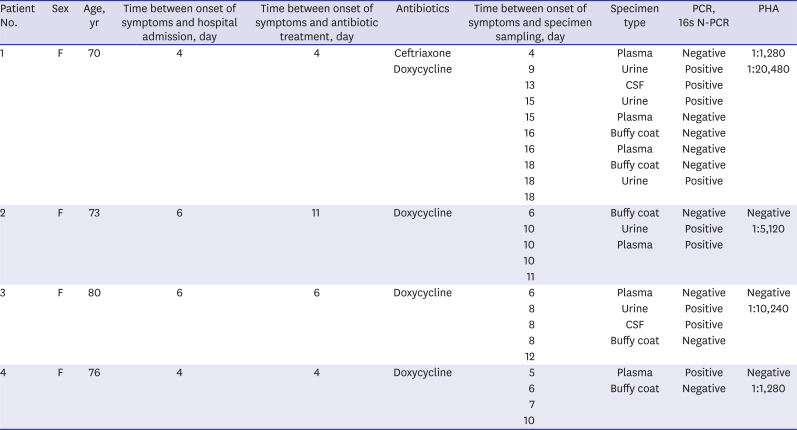
A 70-year-old Korean female was referred to our hospital because of massive hemoptysis and modified Medical Research Council (mMRC) grade 4 dyspnea on September 19, 2008. Three days before this admission, she had abdominal pain, fever, chills, myalgia, and nonproductive cough. She had a history of hypertension. She was a farmer and worked mainly in the fields. Upon physical examination, she had low blood pressure (80/60 mmHg), tachypnea (30 breaths/min), tachycardia (120 beats/min), inspiratory crackle on both lung fields, and conjunctival suffusion. Chest radiography revealed bilateral areas of diffuse infiltrative opacification. A computed tomography (CT) scan of the chest showed diffuse ground-glass opacities (GGO), interlobular septal thickening, and a moderate amount of pleural effusion in both lungs. Laboratory findings showed white blood cells of 9,980/µL (normal range, 4,000–8,000/µL), hemoglobin of 8.1 g/dL (normal range, 12–16 g/dL), platelets of 163 × 103/µL (normal range, 150–400 × 103/µL), and creatinine of 1.47 mg/dL (normal range, 0.6–1.4 mg/dL). Arterial blood gases revealed a pH of 7.406, pCO2 of 26 mmHg, and pO2 of 49 mmHg with FiO2 of 90%. The patient was immediately transferred to the intensive care unit and intubated due to acute respiratory failure. She was treated empirically with oral doxycycline and intravenous ceftriaxone. At 4 and 13 days after symptom onset, the antibody titers using passive hemagglutination assay (PHA) were 1:1,280 and 1:20,480, respectively. N-PCR for Leptospira spp. showed a positive result from urine 9 and 15 days after symptom onset, but N-PCR with the plasma and buffy coat 16 and 18 days after symptom onset was negative. Six days after treatment, the respiratory symptoms and hypoxemia had resolved, and extubation was performed. The patient was discharged home 18 days after symptom onset. At that time, N-PCR using a urine sample was positive.
Case 2
A 73-year-old Korean female was admitted to the emergency room with a 6-day history of headache, myalgia, fever, and chills on August 9, 2007. She had no medical history. She reported that she had worked in the fields 10 days before the admission. She presented with fever (37.7°C). Laboratory results showed white blood cells of 13,480/µL, hemoglobin of 10.4 g/dL, and platelets of 96 × 103/µL. A lumbar puncture test was normal. The antibody titer using PHA was negative at admission. Supportive treatment was performed considering that the fever was due to a viral infection. However, the antibody titer 10 days after symptom onset was 1:5,120. At the same time, Leptospira spp. N-PCR with urine was positive, and also, N-PCR with plasma showed a positive result 11 days after symptom onset. Thereafter, treatment with doxycycline was started. She was discharged home after a 14-day course of doxycycline.
Case 3
An 80-year-old Korean female, who had no previous medical history, was admitted with a headache on September 6, 2007. Six days before admission, she had myalgia, chills, fever, and a sore throat. Upon physical examination, she had a high-grade fever (38.6°C). The laboratory results showed white blood cells of 2,890/µL, hemoglobin of 13.2 g/dL, and platelets of 112 × 103/µL. Urinalysis showed 1+ protein, 4+ blood, with many red blood cells, and 0–1 white blood cells/high-power field. A lumbar puncture test was performed. The CSF showed a white blood cell count of 57/µL, neutrophils of 83%, lymphocytes of 13%, protein of 42 mg/dL, and glucose of 71.9 mg/dL. The antibody titer at admission was negative. N-PCR for Leptospira spp. with plasma showed a negative result, but N-PCR with urine and CSF was positive 8 days after symptom onset. The antibody titer was 1:10,240 12 days after symptom onset. She was discharged home with a 14-day course of doxycycline.
Case 4
A 76-year-old Korean female was referred to our hospital because of an altered mental status on August 30, 2007. Four days before the admission, she presented with general weakness and myalgia. She had no medical history. Upon physical examination, she had stuporous mentation, low blood pressure (80/50 mmHg), tachypnea (28 breaths/min), tachycardia (108 beats/min), and an inspiratory crackle on both lung fields. The laboratory results showed white blood cells of 10,190/µL, hemoglobin of 9.8 g/dL, platelets of 83 × 103/µL, and creatinine of 2.31 mg/dL. Urinalysis showed 1+ protein, 5+ blood, with many red blood cells, and 5–9 white blood cells/high-power field. Chest radiography revealed bilateral areas of diffuse infiltrative opacification. After admission, the patient was intubated due to acute respiratory distress. A CT scan of the chest showed bilateral areas of ground-glass attenuation with superimposed interlobular septal thickening. The antibody titer was negative 5 days after symptom onset, but N-PCR with a plasma sample was positive 6 days after symptom onset. The antibody titer was 1:1,280 10 days after symptom onset. After 5 days of treatment with doxycycline, the patient was extubated. Shortly thereafter, she went into acute respiratory distress and died, although mechanical ventilation was applied.
Ethics statement
The study without informed consent was approved by the Institutional Review Board (IRB) of Chosun University Hospital (IRB No. 2018-04-007).
DISCUSSION
We described 4 cases of patients diagnosed with leptospirosis using N-PCR that targeted the 16S rRNA gene. The clinical characteristics of the patients are summarized in
Table 1. N-PCR permitted the early detection of leptospirosis, and it was also useful with specimen types (urine or CSF) other than the plasma or buffy coat. The plasma may be better than the buffy coat as a blood specimen type for N-PCR.
Table 1
Clinical characteristics of 4 leptospirosis patients
|
Patient No. |
Sex |
Age, yr |
Time between onset of symptoms and hospital admission, day |
Time between onset of symptoms and antibiotic treatment, day |
Antibiotics |
Time between onset of symptoms and specimen sampling, day |
Specimen type |
PCR,16s N-PCR |
PHA |
|
1 |
F |
70 |
4 |
4 |
Ceftriaxone |
4 |
Plasma |
Negative |
1:1,280 |
|
Doxycycline |
9 |
Urine |
Positive |
1:20,480 |
|
13 |
CSF |
Positive |
|
|
15 |
Urine |
Positive |
|
|
15 |
Plasma |
Negative |
|
|
16 |
Buffy coat |
Negative |
|
|
16 |
Plasma |
Negative |
|
|
18 |
Buffy coat |
Negative |
|
|
18 |
Urine |
Positive |
|
|
18 |
|
|
|
|
2 |
F |
73 |
6 |
11 |
Doxycycline |
6 |
Buffy coat |
Negative |
Negative |
|
10 |
Urine |
Positive |
1:5,120 |
|
10 |
Plasma |
Positive |
|
|
10 |
|
|
|
|
11 |
|
|
|
|
3 |
F |
80 |
6 |
6 |
Doxycycline |
6 |
Plasma |
Negative |
Negative |
|
8 |
Urine |
Positive |
1:10,240 |
|
8 |
CSF |
Positive |
|
|
8 |
Buffy coat |
Negative |
|
|
12 |
|
|
|
|
4 |
F |
76 |
4 |
4 |
Doxycycline |
5 |
Plasma |
Positive |
Negative |
|
6 |
Buffy coat |
Negative |
1:1,280 |
|
7 |
|
|
|
|
10 |
|
|
|
We conducted N-PCR in the following procedures. Total DNA from human specimens was prepared using QIAamp DNA Mini kits (QIAGEN, Germantown, MD, USA), according to the manufacturer's instructions. The primers were designed from alignments of available Leptospira spp. 16S rRNA sequences, obtained from the GenBank nucleotide sequence database. The designed primers were the external primer L2-exF583 (5′-gaaactatgtgtctggagtttgg-3′), L2-exR1211 (5′-cctcgcgagttggctacc-3′), internal primer L2-inF780 (5′-gggggttttaaccctcagtaa-3′), and L2-inR987 (5′-cccgaaggctcatgtatctc-3′). N-PCR was performed using the AccuPower® PCR PreMix (Bioneer, Daejeon, Korea) targeting a 200 bp fragment of the 16S rRNA gene. PCR amplicons were loaded on a 1.5% agarose gel and then visualized by a UV illuminator.
In the acute or septicemic phase, DNA- or antigen-based tests can be used to detect the presence of the leptospiral antigen and DNA in the blood.
15 The laboratory diagnosis of leptospirosis varies and includes serological tests, molecular tests, and culture. Isolation of a pathogenic
Leptospira species from a sterile site may take several weeks to obtain results with low sensitivity (5%–50%).
6 The leptospiremia is usually finished by the end of the first week after infection. Samples from CSF may be cultured during the first week of illness. Urine can be cultured from the beginning of the second week after the onset of symptoms.
1
Serological tests include the MAT, immunoglobulin M (IgM) enzyme-linked immunosorbent assay (ELISA), indirect hemagglutination assay (IHA) including PHA, immunochromatography test, etc. MAT is considered the gold standard test for the diagnosis of leptospirosis, as it detects both the IgM and IgG class of agglutinating antibodies,
7 but MAT may give false negative results in the early acute phase of the disease, because the IgM antibody titers are usually low in the blood approximately 5-7 days after the onset of symptoms.
125 Bajani et al.
8 reported that the sensitivity of IHA was 38.5% for acute-phase sera, 67.2% for convalescent-phase sera and the specificity was 95.8%. The antibody titer using PHA was negative in 3 of the 4 cases at admission. A reliable and valid, rapid diagnostic test for leptospirosis that permits the diagnosis and treatment of leptospirosis in the early course of infection is needed. Niloofa et al.
9 reported that the IgM ELISA (Virion/Serion) and immunochromatography test (Leptocheck-WB Test; Zephyr Biomedicals, Verna, India) are suitable for an early and definitive diagnosis or as a screening test for acute leptospirosis, due to the high sensitivity and reasonable specificity during the acute phase of illness.
Molecular tests for the diagnosis of leptospirosis, such as conventional and real-time PCR, reverse-transcription PCR, and isothermal amplification methods, have been developed. Targeting housekeeping genes that are included in all species of
Leptospira (16S
rrs,
secY, or
gyrB) or pathogenic species-specific genes (
lipL32,
lfb1, or
lig) have been used as targets for PCR-based diagnosis.
210111213 The sensitivity and specificity of PCR has been reported as 52.7% and 97.2%, respectively, compared to MAT (49.8% and 98.8%, respectively).
3 PCR for the detection of pathogenic
Leptospira spp. DNA has an advantage over serologic tests, especially in the acute phase. In our 4 cases,
Leptospira DNA was detected in urine, plasma, or CSF in the early phase of leptospirosis by N-PCR, but not from the buffy coat.
Leptospira DNA has been amplified from serum, urine, aqueous humor, CSF, and a number of tissues obtained at autopsy.
110 In cases 2 and 4, N-PCR for
Leptospira spp. from plasma was positive, but the sample from the buffy coat was negative at that time. Kositanont et al.
14 reported that the buffy coat was superior to plasma and serum. In their report, leptospiral DNA was detected by PCR in all 7 buffy coat preparations from laboratory-confirmed leptospirosis cases, whereas leptospiral DNA was detected in only 3 of the 7 plasma or serum samples. However, Wood et al.
15 reported that there was no difference in sensitivity between serum and buffy coat by quantitative PCR (qPCR), and serum was more suitable than buffy coat due to qPCR inhibition.
The sensitivity of PCR for
Leptospira DNA detection from blood declines over the course of the disease.
216 Case 4 showed that N-PCR with plasma was positive 6 days after symptom onset. However, N-PCR with urine and/or CSF was positive during the second week after symptom onset, except for the samples from the plasma and buffy coat in cases 1 and 3. Additionally, case 1 showed that N-PCR from a urine sample was positive 18 days after symptom onset.
Leptospira DNA in urine and CSF can be detected for a longer duration than from blood or serum. Bal et al.
17 detected
Leptospira in 26 of the 29 urine samples from patients with leptospirosis by PCR, for which 7 of the urine samples were obtained before day 8 of illness and 19 of the urine samples were obtained after day 8 of illness. Waggoner et al.
11 reported 2 cases of leptospirosis, in which
Leptospira was detected in the CSF at a bacterial load 5- to 10-fold higher than that in the plasma by qPCR. More studies are needed conventional analysis of diagnostic accuracy using molecular techniques compared with the reference methods (MAT and culture) according to the acute- and convalescent-phase and the specimen types.
Our study has limitations. First, it is a case series from a single tertiary center, with data collected from patients' medical records, making it susceptible to bias in data selection and analysis. Second, all patients in the case series were not tested at the same time between the onset of symptoms and specimen sampling, and the types of specimens obtained were also different. Third, we diagnosed leptospirosis by N-PCR and PHA, not reference methods.
In conclusion, we report 4 cases of leptospirosis that were diagnosed by N-PCR that targeted the 16S rRNA gene from urine, plasma, or CSF samples. Detection of Leptospira DNA by PCR from urine and CSF, in addition to plasma, may be helpful to confirm the diagnosis. Further research requires an effectiveness analysis of the molecular tests using urine and CSF together with the serum, plasma, buffy coat, or whole blood for the diagnosis of leptospirosis.