Sapovirus belongs to the
Caliciviridae family, such as norovirus and is genetically classified into five groups (GI-GV). GI, GII, GIV and GV are known to cause human infection, except for GIII which causes infection in pigs.
12 Transmission of sapovirus occurs via fecal oral route from contaminated water and food, and person-to-person spread. Also, the most common symptoms of sapovirus infection are similar to norovirus infection, such as vomiting, diarrhea, nausea, chills, and fever. The incidence of gastroenteritis associated with sapovirus has been reported throughout the year in all age groups in a variety of settings, such as in childcare centers, colleges, nursing homes, hospitals, restaurants, cruises and schools.
3456 Sapovirus shows a low prevalence (below 1%–2%), according to EnterNet-Korea, which is the Enteric Pathogens Active Surveillance Network-Korea. In this study, we report that sapovirus can be a causative agent of gastroenteritis outbreak in elementary school students and characterize molecular epidemics of sapovirus associated with this outbreak.
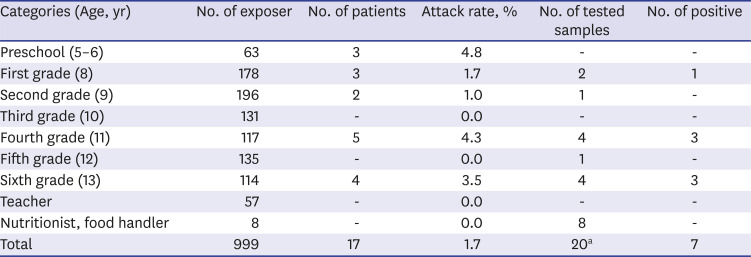
On October 4, 2018, there was an outbreak of acute gastroenteritis in one elementary school in Gyeonggi Province. Epidemiologic studies were conducted in a retrospective cohort approach. The cases were defined as having diarrhea three times a day or vomiting three or more times a day, and interviews were conducted using standardized questionnaires. The total number of exposed people was 999, including kindergarten and elementary school students, school staff, and cook workers. Among them, 17 respondents were found to meet the definition. The respondents were divided as follows: three kindergartens, three in the first grade, two in the second grade, five in the fourth grade, and four in the sixth grade (
Table 1). The first clinical symptoms were reported at dawn on October 4, and the last was reported at dawn on October 5; occurring for about 24 hours. The most common symptom was reported on the afternoon of October 4th (8 patients) (
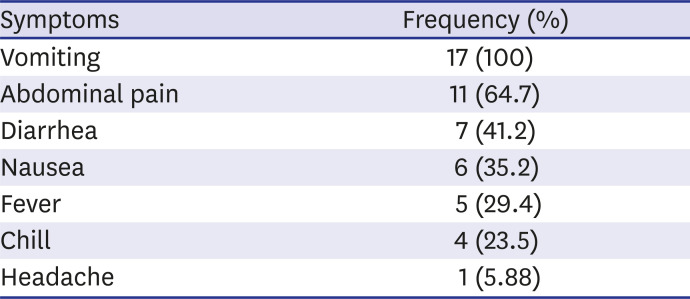
Fig. 1). The main clinical symptoms were vomiting (17 patients, 100%), abdominal pain (11 patients, 64.7%) and diarrhea (7 patients, 41.2%) (
Table 2).
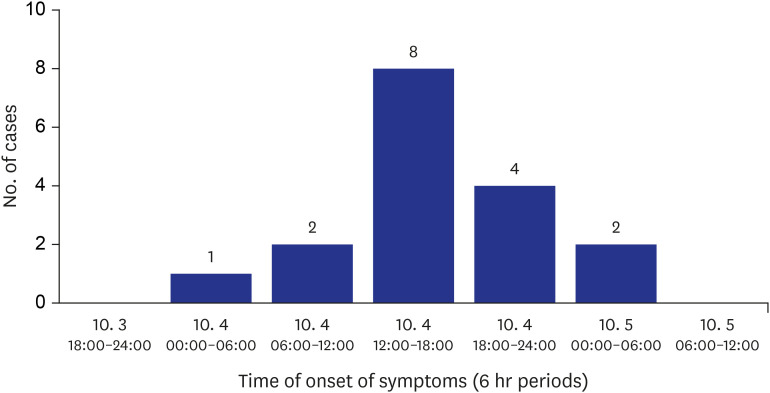 | Fig. 1Epidemic curve of the outbreak. Clinical symptoms began on October 4 and terminated on October 5. Individual cases experienced general clinical symptoms, which included vomiting, abdominal pain, and diarrhea. 
|
Table 1
Attack rates and the results of stool samples for sapovirus
|
Categories (Age, yr) |
No. of exposer |
No. of patients |
Attack rate, % |
No. of tested samples |
No. of positive |
|
Preschool (5–6) |
63 |
3 |
4.8 |
- |
- |
|
First grade (8) |
178 |
3 |
1.7 |
2 |
1 |
|
Second grade (9) |
196 |
2 |
1.0 |
1 |
- |
|
Third grade (10) |
131 |
- |
0.0 |
- |
- |
|
Fourth grade (11) |
117 |
5 |
4.3 |
4 |
3 |
|
Fifth grade (12) |
135 |
- |
0.0 |
1 |
- |
|
Sixth grade (13) |
114 |
4 |
3.5 |
4 |
3 |
|
Teacher |
57 |
- |
0.0 |
- |
- |
|
Nutritionist, food handler |
8 |
- |
0.0 |
8 |
- |
|
Total |
999 |
17 |
1.7 |
20a
|
7 |

Table 2
Symptom-wise distribution of cases (n = 17)
|
Symptoms |
Frequency (%) |
|
Vomiting |
17 (100) |
|
Abdominal pain |
11 (64.7) |
|
Diarrhea |
7 (41.2) |
|
Nausea |
6 (35.2) |
|
Fever |
5 (29.4) |
|
Chill |
4 (23.5) |
|
Headache |
1 (5.88) |

In order to identify causative pathogens, human and environmental specimens were collected and examined. Microbiological tests were performed at the Gyeonggi-do Institute of Health and Environment, following the laboratory diagnosis guidelines of the Korea Center for Laboratory Control of Infectious Diseases (KCDC) and the Ministry of Food and Drug Service (MFDS). The molecular epidemiological investigation was performed at the Division of Viral diseases, KCDC. A total of 20 fecal samples were collected by rectal smear method, including 11 of 17 patients who fulfilled the definition, 8 persons who worked as cooks, and 1 patient who had other symptoms. The pathogens to be tested included 10 species of bacteria (Shigella spp, Salmonella spp, pathogenic E. coli (EHEC, ETEC, EPEC, EIEC etc.), Vibrio spp, Campylobacter jejuni, Staphylococcus aureus, Listeria monocytogenes, Clostridium perfringens, Yersinia enterocolitica, Bacillus cereus) and 5 species of viruses (Group A rotavirus, norovirus, enteric adenovirus, astrovirus, sapovirus). The environmental samples were basically sampled from preservated foods, drinking water, cooking utensils, and cooking water. Specimens were also sampled from door knobs, handrails, corridors in front of the office, and vomiting areas in places where guests spent much time. In environmental samples, only 10 species of bacteria were tested.
Bacterial tests were confirmed by biochemical tests on the specific colonies formed after cultivation in the selective medium. For virus detection, the supernatant was used in the experiment after centrifugation of the feces dilution. Group A rotavirus and intestinal adenovirus were confirmed by EIA, and norovirus, astrovirus and sapovirus were confirmed by genetic testing. To briefly describe the sapovirus assay, the pre-treated supernatant was extracted with a commercially available nucleic acid extraction kit (NucleoMag 96® Virus kit, Macherey-Nagel, Dueren, Germany) according to the manufacturer’s instructions. Sapovirus was detected by reverse transcriptase-polymerase chain reaction (RT-PCR) using a commercial one-step premix kit (iMOD-001TD, SnC, Seoul, Korea) with the following conditions: 48°C, 40 minutes; 94°C, 15 minutes; 94°C, 30 seconds; 58°C, 30 seconds; 72°C, 60 seconds for 35 cycles; and 72°C 7 minutes. The oligo nucleotide sequence was performed using forward primer SV-F21 (5′-ANT AGT GTT TGA RAT GGA GGG-3′) and reverse primer SV-R1 (5′-CWG GTG AMA CMC CAT TKT CCA T-3′) to amplify the ORF1 capsid region. In the genetic test, only seven samples of human specimens were found to have sapovirus, with 1 in grade 1, 3 in grade 4, and 3 in grade 6 (
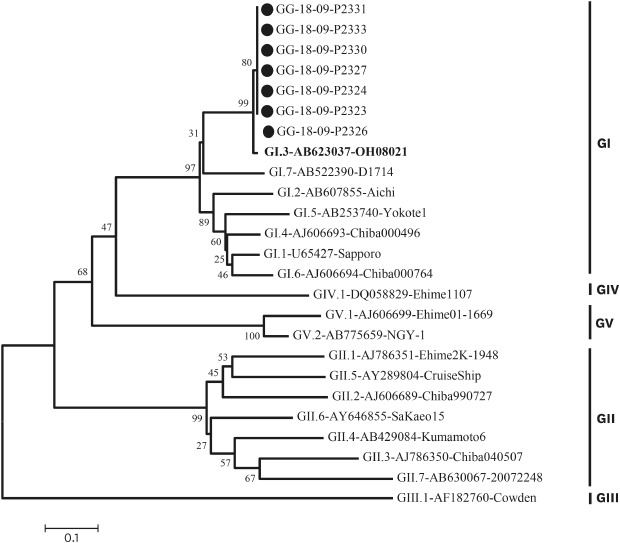
Table 1). Nothing was confirmed in environmental samples. Molecular epidemiological analyses were performed on specimens identified as positive for sapovirus. The nucleotide sequence was analyzed using a 3730xl automated sequencer (Applied Bio Systems, Foster City, CA, USA). The genotype was then determined by sequencing followed by BLAST searching of the GenBank database and alignment using the Bioedit 7.0 program. We used the neighbor-joining method (Kimura 2-parameter model with 1,000 bootstrap replicates) of the MEGA 6 analysis program, and all 7 cases formed the same cluster as sapovirus GI.3 (
Fig. 2).
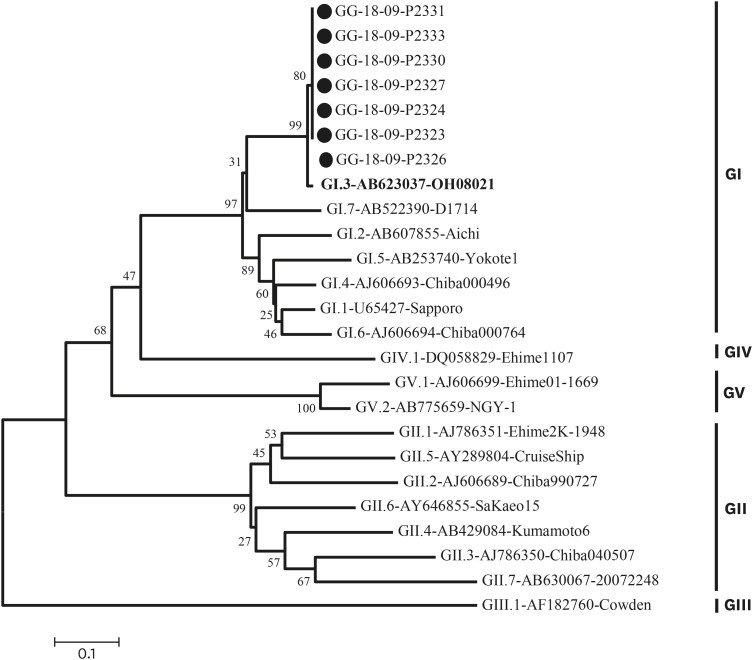 | Fig. 2Phylogenetic analysis of sapovirus detected in the outbreak, based on 720 bp of the capsid region (ORF1) sequences. The phylogenetic tree was generated using the neighbor-joining method with distance calculation by Kimura-2-parameter correction implemented in the MEGA6 software. The numbers in the branches indicate the bootstrap of 1,000 replicated values. Reference strains of sapovirus genotype were registered in GenBank under the accession number indicated in the text. Outbreak samples are marked with black circles (●). 
|
One of the human caliciviruses, sapovirus has emerged as an important cause of viral gastroenteritis in people of all age groups and is the cause of outbreaks. Sapovirus associated outbreak has been increasing globally. We described an outbreak of gastroenteritis associated with sapovirus in one elementary school in Gyeonggi Province, Korea. In this study, symptoms due to sapovirus infection were observed in 1.7% (17/999) of the subjects (
Table 1). Common symptoms of sapovirus infection are known to be associated with diarrhea, vomiting, abdominal pain, fever, and chills.
2 Shuzo et al.
7 and Johansson et al.
8 reported that diarrhea is more common than vomiting in adults. In the case of norovirus, more than 50% of the cases have vomiting symptoms in children, which may be a characteristic of viral infection belonging to Calicivirus.
9 In this case, all of the respondents (17 patients, 100%) showed vomiting symptoms and only 7 patients (41.2%) showed diarrhea, so the main symptom of sapovirus infection in children is vomiting, similar to norovirus (
Table 2).
In this outbreak there were 7 sapovirus-positive samples from students in multiple grade levels. Epidemiologic studies have shown that volunteers had no connection through family relationships or contact through social activities such as academies and clubs. Considering that the epidemic curve has been reported in most cases to occur within 24 hours, it is estimated that a single exposure by water or food was more likely to have occurred than person-to-person transmission. However, we were not able to determine the infectious agent because the virus was not detected in cooks or environmental samples. Moreover, phylogenetic analysis showed that the strain that shows the most resemblance is the GI.3 strain (strain sapovirus Hu/OH08021/JP: Genbank accession number AB0623037), which was isolated in Japan in 2008. The epidemic strain in Korea was GI.1 type which was confirmed by a domestic acute diarrheal laboratory surveillance system. Analysis of genotypes is important for identifying currently circulating sapoviruses. The most common sapovirus genogroups are GI and GII.
10 Genotype I.1 is the most common genotype around the globe, and GI.2 has been increasingly detected in many European countries,
11 and also in northern Africa.
12 In Korea, as previously reported, the most common genotype is GI.1, but the genotype of the outbreak was GI.3, which is not a domestic genotype.
Although there are limitations, it is important to report the first infection in Korea due to sapovirus infection. The main clinical symptom in children with sapovirus infection was vomiting, not diarrhea, and the potential for infection by rare genotypes is also an important finding of this case study.







 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download