INTRODUCTION
Oxidative stress, inflammation, and immune system activation are known to be closely related to metabolic disorders such as diabetes, obesity, and associated cardiovascular complications.
1) When atherosclerosis develops, metabolic stress may significantly affect the inflammatory response of endothelial cells, monocytes, and macrophages.
2) Although the mechanism is not completely understood, diverse types of oxidized low-density lipoproteins (LDLs) are have also been shown to contribute to steatohepatitis. Thus, intervention or inhibition of certain metabolic pathways is being considered as a therapeutic option against inflammatory disorders.
3)
Saturated fatty acids and oxidized LDL commonly coexist and contribute to vascular inflammation. Saturated fatty acids can induce inflammatory response via the toll-like receptor (TLR) 4 pathways, and can affect lipopolysaccharide (LPS) production via modification of gut microbiota. Subsequent endotoxemia may trigger oxidative stress and result in the production of oxidized LDL, a major form of atherogenic lipids.
4) We chose palmitate (PA) in this study, as it is a major form of saturated fatty acid in human circulation, and it is a player in metabolically pathologic conditions like insulin resistance.
5) Several studies have reported metabolic inflammation caused by PA.
5)6)
The purpose of this study was to evaluate proinflammatory effects and to identify genes regulated by metabolic stimuli, saturated fatty acids or modified LDL. Specifically, we aimed to examine the effects of PA and minimally modified LDL (mmLDL) on LPS-primed macrophages with pivotal roles in chronic inflammatory conditions, such as atherosclerosis. We also investigated the genes regulated in this process and validated the expression of some of them. Our targets included NLRP3, an essential mediator of the interleukin (IL)-1 pathway, which is currently being actively investigated. The IL-1 pathway upregulated in vascular inflammation, is one of key pathways suggested as a target for anti-inflammatory therapy. For example, the CANTOS clinical trial showed that an IL-1β antagonist reduced cardiovascular risk.
7) Building on our previous research on the combined effects of PA and mmLDL,
8) we sought to further investigate the individual effects of these 2 agents and genes associated with the observed effects in the current study.
Go to :

METHODS
Macrophages and other reagents
Dulbecco's modified Eagle's medium (DMEM), penicillin-streptomycin, fetal bovine serum (FBS) and Dulbecco's phosphate-buffered saline were purchased from Gibco (Grand Island, NY, USA). The murine macrophage-like cell line J774A.1 (abbreviated as J774) was provided by Yury I Miller in University of California, San Diego, USA. LPS and PA were purchased from Sigma-Aldrich (St. Louis, MO, USA) for macrophage stimulation. U0126, an extracellular signal-regulated kinase (ERK) inhibitor, was purchased from Cell Signaling Technology (Danvers, MA, USA). It was dissolved in dimethyl sulfoxide (DMSO), and a stock solution of 10 mM was prepared before experiments. TAK-242, a TLR4 inhibitor, was purchased from InvivoGen (San Diego, CA, USA). PA was dissolved in 0.1 M 70% ethanol and a control solution containing ethanol was also prepared. TAK-242 was dissolved in DMSO. Stock solutions of 30 mM PA and 10 mM TAK-242 were prepared before the experiment. LDL (density, 1.019−1.063 g/mL) was isolated from the plasma of healthy donors using sequential ultracentrifugation.
9) To produce mmLDL, 50 µg/mL LDL was incubated in serum-free DMEM for 18 hours with murine fibroblast cells overexpressing human 15-lipoxygenase.
8) mmLDL has been characterized in detail previously.
10) Preparations of PA and mmLDL were assessed for LPS contamination using the Limulus amebocyte lysate assay (Lonza, Basel, Switzerland). Endotoxin levels were < 0.05 EU/mL in all experiments. We chose 100 µM PA
8)11)12) and 50 µM/mL mmLDL
8)13) for the experiment as these doses have frequently been used to test effects of PA or mmLDL on macrophages in previous studies.
Cell culture and treatment
J774 cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin- streptomycin (Gibco, Carlsbad, CA, USA) at 37°C in a 5% CO2 environment. Prior to stimulation, the cells were maintained in serum-poor (0.5% FBS) medium for 6 hours. For protein expression analysis, J774 cells were plated (5 × 105 cells/well) on 60-mm 6-well plates and incubated overnight. The cells were then either not treated or treated with 100 µM PA for 18 hours before being stimulated with 3 ng/mL LPS. Otherwise, the cells were treated with 3 ng/mL LPS with or without 50 µg/mL mmLDL for 6 hours. After 30 minutes, the cells were harvested for immunoblotting. The supernatant was collected 6 hours post-treatment with LPS with or without mmLDL, and then analyzed using enzyme-linked immunosorbent assay (ELISA). For quantitative polymerase chain reaction (qPCR) analysis, cells were harvested 6 hours post-treatment with LPS with or without mmLDL. All experiments were conducted in biological triplicates.
Enzyme-linked immunosorbent assay
The cell culture supernatant was collected and the amounts of secreted IL-6, IL-1β, or tumor necrosis factor (TNF)-α were quantified using ELISA kits (R & D Systems, Minneapolis, MN, USA) according to the manufacturer's protocols. Technical duplicates of all samples were used.
Immunoblot analysis
Cells were harvested and solubilized in radioimmunoprecipitation assay buffer (Biosesang, Seongnam, Korea) containing a protease inhibitor cocktail (Roche, Inc., Indianapolis, IN, USA). We determined the protein concentration using a bicinchoninic acid protein assay (Pierce Biotechnology, Inc., Rockford, IL, USA). Cell lysates were resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membrane was then incubated at room temperature for 1 hour with 5% bovine serum albumin in Tris-buffered saline (TBS) solution with Tween-20 and with a primary antibody overnight at 4°C. On the following day, the membranes were washed with TBS and incubated with a horse radish peroxidase-conjugated secondary antibody at room temperature. The immunoreactive bands were visualized using the enhanced chemiluminescence kit (EMD Millipore).
RNA extraction and real-time polymerase chain reaction
RNA was extracted from cells using a Ribospin RNA extraction kit (GeneAll, Seoul, Korea) according to the manufacturer's protocol. RNA concentration and purity were assessed using a NanoDrop spectrophotometer. One microgram of RNA was then used for complementary DNA (cDNA) synthesis with a QuantiTect reverse transcription kit (Qiagen, Venlo, Netherlands). Real-time polymerase chain reaction (PCR) was performed using the SYBR dye system and Step One Plus real-time PCR machine (Applied Biosystems, Foster City, CA, USA). Gene expression analysis was performed using the LightCycler software based on cycle threshold values and is shown as fold change of expression. Messenger RNA (mRNA) levels were normalized to glyceraldehyde 3-phosphate dehydrogenase mRNA. All results were derived from technical duplicates.
RNA sequencing and analysis
RNA purity was determined using a NanoDrop ND1000 spectrophotometer. Total RNA sequencing libraries were prepared according to the manufacturer's instructions using a Truseq stranded total RNA sample prep kit (Illumina, San Diego, CA, USA). One microgram of total RNA was further treated for ribosomal RNA (rRNA) depletion using Ribo-zero rRNA removal kit (human/mouse/rat) (Illumina). Then, the rRNA-depleted total RNA was fragmented into small pieces at 94°C for 8 minutes. The cleaved RNA fragments were copied to first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA polymerase I and RNase H. Finally, one 'A' base was added, and the adapter was subsequently ligated. The products were purified and enriched using PCR to create the final cDNA library. After real-time PCR, index-tagged libraries were combined in equimolar ratio. RNA sequencing was performed using the Illumina NextSeq 500 platform according to the manufacturer's recommended protocol.
The reads were aligned with the gene expression values using Cufflinks (fragments per kilobase of transcript per million mapped reads). The differentially expressed genes of the vehicle treatment group were compared to that of the PA or mmLDL treatment group using Cuffdiff. Upregulated or downregulated genes with q-value <0.05 and 2-fold change were identified.
Small interfering RNA transfection
Small interfering RNAs (siRNAs) against Csf3r and Edn1 and the scrambled siRNA were purchased from Mbiotech (Hanam, Korea). For gene silencing, J774 cells were treated with 100 nM siRNA using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA). After 24 hours, the media with siRNA were removed and stimulants were treated for each experiment.
Statistical analysis
All data are presented as the mean±standard error of the mean. We performed t-test for comparison between groups. p<0.05 (2-sided) was considered statistically significant. We used the software package Prism 5.0 for all data analyses (GraphPad Software Inc., San Diego, CA, USA).
Go to :

DISCUSSION
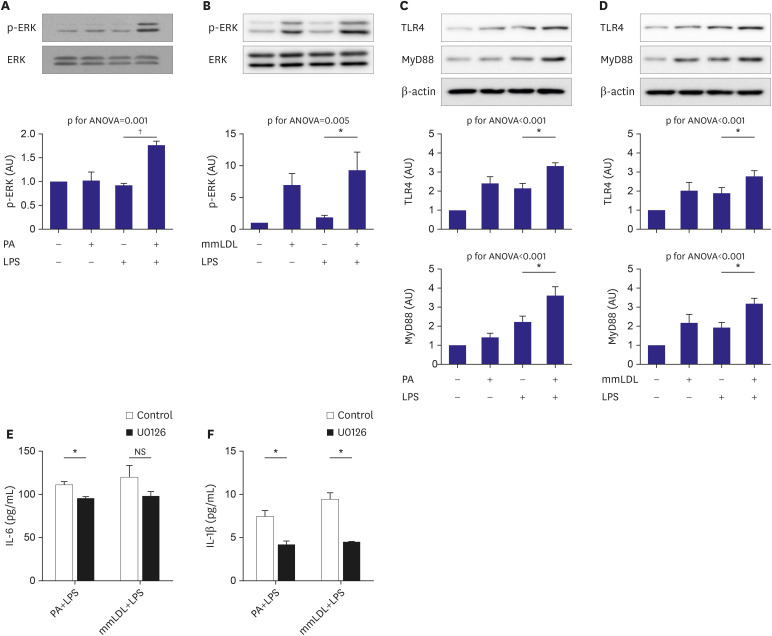
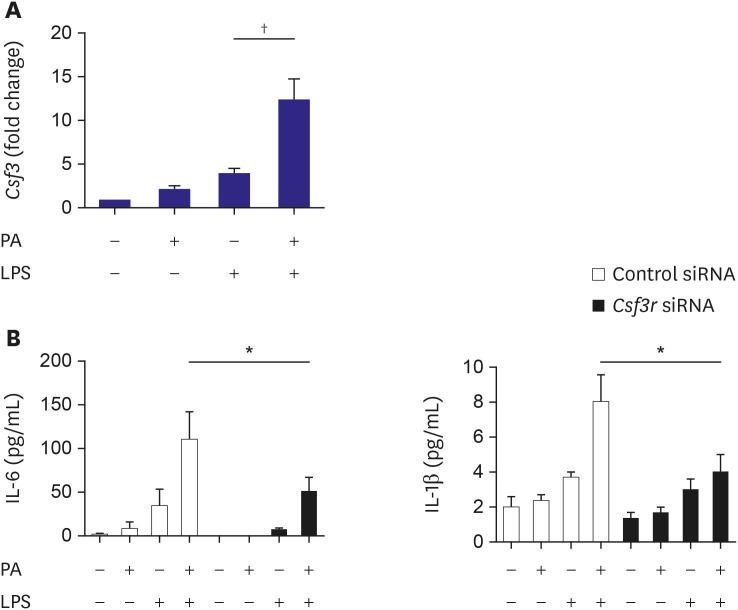
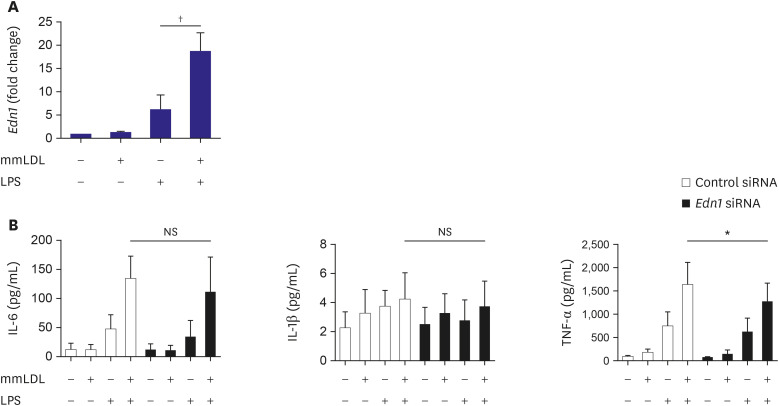
The findings of our study include: 1) PA or mmLDL promoted inflammatory chemokine secretion in LPS-stimulated macrophages. 2) This was accompanied by the activation of the ERK MAP kinase pathway. 3) Dozens of PA and mmLDL-regulated genes were identified, which included Csf3 and End1, respectively. 4) Nlrp3 was also found to be activated by these 2 agents. 5) The effect of silencing either Csf3 or Edn1 appeared to be modest, whereas TLR4 inhibition reduced a large proportion of macrophage activation by PA or mmLDL. In this study, we demonstrated the effect of the proinflammatory metabolic milieu on macrophages and the associated molecular pathways.
We demonstrated, for the first time, that mmLDL can upregulate
Nlrp3. Although the TLR4 pathway appeared to contribute the most to this process, evidence on the role of other genes was also provided. The strength of the current study also relies on the identification of genes regulated by PA or mmLDL. These can be suggested as novel therapeutic targets against poor metabolic conditions. It has been reported that PA could promote insulin resistance and inflammation via TLR4-dependent and some other pathways.
17) However, we are the first to provide data demonstrating that
Csf3 is upregulated by PA. Conversely, the pathways on which oxidized LDL works have been substantially investigated and understood, particularly, oxidized LDL's interactions with CD36 and its influence on inflammasomes.
18) Remarkably, however, we revealed that mmLDL not containing advanced lipid oxidation products upregulates
Nlrp3.
The NLRP3 inflammasome is a well-known signaling complex involved in innate immunity and is a key mediator of the production of IL-1 family of cytokines. It is activated by various endogenous signals during the progression of atherosclerosis, which include pathogen-associated molecular patterns, danger-associated molecular patterns, and oxidized LDL.
19) Free fatty acids such as PA have been reported to promote NLRP3 production via the pattern recognition receptor.
19) Oxidized LDL is known to be involved in the formation of the NLRP3 inflammosome.
19)20) This effect of oxidized LDL can also result in disorders not only in cardiovascular organs. Uptake of oxidized LDL by Kupffer cells has been reported to induce inflammasome activation, impairment of autophagy and hepatic inflammation and non-alcoholic steatoheaptitis. In addition, activated inflammasome and promoted apoptosis of hepatocytes are known to induce inflammation. Furthermore, oxidized LDL can activate hepatic stellate cells and leads to pro-fibrotic response.
21) However, for the first time, we showed that mmLDL upregulated
Nlrp3 expression.
Studies have suggested that PA promotes inflammation via multiple pathways, and TLR2 and TLR4 are considered the major receptors mediating this effect. However, alternative mediators such as diacylglycerol, ceramide, and lysophosphatidylcholine also contribute to intracellular inflammation by PA. Hence, controlling the response to PA by inhibiting a single pathway may be challenging.
22) Reports have shown that mmLDL promotes inflammation via TLR4.
23) Choi et al.
24) showed that oxidized cholesteryl ester of mmLDL induces secretion of inflammatory cytokines, which is mediated by TLR4 and spleen tyrosine kinase. In our study, a TLR4 inhibitor attenuated a large proportion of IL-1β secretion. Thus, we confirmed that this pathway can be a target for efficiently controlling macrophage activation with PA or mmLDL. We confirmed that PA or mmLDL activated ERK MAPK and upregulated TLR4 in LPS-stimulated macrophages. This finding was validated by using ERK inhibitor. However, because the activation of ERK by these agents was previously shown and was not the main purpose of the current study, we did not further explore ERK-related issues.
In the present study, we observed that silencing of 2 genes,
Csf3 and
Edn1, modestly but significantly reduced the cellular effect of PA and mmLDL, respectively.
Csf3 function has been reported to be dependent on TLR4.
8) This gene is associated with inflammation and is upregulated in the epicardial adipose tissue of patients with diabetes and coronary artery disease. This suggests that
Csf3 expression may be related to metabolism and atherosclerosis.
25) Furthermore,
Csf3 was reported to be one of the differentially expressed genes during the dedifferentiation of human smooth muscle cell phenotype, indicating its possible relationship with atherogenesis.
26) Upregulation of
Csf3 by PA in our study implied that
Csf3-related pathways may contribute to the proinflammatory and proatherogenic activities of PA.
We observed that
Edn1 was upregulated by mmLDL. A previous human genetic study has revealed the association of
Edn1 with fibromuscular dysplasia or coronary artery disease.
27)
Edn1 is one of the transcriptionally regulated genes in macrophage response.
28) In
Cd73-deficient mice,
Edn1 was found to be upregulated and atherosclerosis was promoted.
29) The extent of mmLDL-mediated upregulation of
Edn1 that can contribute to vascular pathology remains to be investigated. The other genes affected by PA or mmLDL include, but are not limited to, the following:
Il1f9,
C1qc, and
Cd93 were upregulated by PA, whereas
Thbs1,
Il1a, and
Slc39a10 were upregulated by mmLDL. However, till date, the role of these genes in cardiovascular health remains uncertain.
In our experiments, LPS frequently showed a greater impact than PA or mmLDL on chemokine secretion or expression of related genes. However, our focus was on PA or mmLDL in LPS-primed macrophages, not to compare any of them. Although an environment with elevated endotoxin, represented by LPS, is of importance,
30) our study sought to investigate the effect of PA and mmLDL, on top of endotoxemia, which are known mediators in poor metabolic conditions.
Our study has a few limitations. The doses of PA or mmLDL used in our study may differ from the concentration found in human tissues or in circulation. However, we referred to doses used in previous studies, and our dosages were within the published ranges. We validated the roles of Csf3 and Edn1, as well as that of TLR4, and observed that the effect of TLR4 inhibition was the most obvious. As TLR4, an upstream receptor, plays a major role in these processes, the feasibility of using other genes that play minor roles in these processes as intervention targets could be uncertain. However, we identified several PA- and mmLDL-affected genes, and multiple relevant genes may possibly act cooperatively. In particular, risk factors other than saturated fatty acids and modified LDL may coexist in metabolic conditions that are prone to vascular diseases. Therefore, an understanding of the diverse risk factors and associated targets will be valuable for translating the results of this study into practice.
In conclusion, we demonstrated that proinflammatory metabolic conditions involving elevation in PA or mmLDL levels promoted macrophage activation via regulation of the expression of various genes such as Nlrp3, Csf3, and Edn1. Although the TLR4 pathway appeared to be the most relevant, validation of the role of other genes provided insights regarding the potential targets for intervention in this environment.
Go to :








 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download