Abstract
Objectives
Oral squamous cell carcinoma (OSCC) is one of the most common types of head and neck cancer. MicroRNAs, as new biomarkers, are recommended for diagnosis and treatment of different types of cancers. Bevacizumab, sold under the trade name Avastin, is a humanized whole monoclonal antibody that targets and blocks VEGF-A (vascular endothelial growth factor A; angiogenesis) and oncogenic signaling pathways.
Materials and Methods
This study comprised 50 cases suffering from OSCC and 50 healthy participants. Peripheral blood samples were collected in glass test tubes, and RNA extraction was started immediately. Expression levels of miR-155, miR-191, and miR-494 biomarkers in the peripheral blood of OSCC-affected individuals and healthy volunteers in vivo were evaluated using real-time PCR. The influence of Avastin on the expression levels of the aforementioned biomarkers in vitro and in the HN5 cell line was also investigated.
Results
Expression levels of miR-155, miR-191, and miR-494 in the peripheral blood of individuals affected by OSCC were higher than in those who were healthy. Moreover, Avastin at a concentration of 400 µM caused a decrease in the expression levels of the three biomarkers and a 1.5-fold, 3.5-fold, and 4-fold increase in apoptosis in the test samples compared to the controls in the HN5 cell line after 24, 48, and 72 hours, respectively.
Go to : 
Oral squamous cell carcinoma (OSCC) is the sixth most common cancer worldwide1. Only 50% of patients survive up to five years because they are diagnosed in advanced stages2. Given the severity and complication of the disease, using a diagnostic tool is necessary for clinical prognosis1. This goal can be achieved by identification of molecular pathways and biomarkers to prognosticate and detect OSCC at early stages.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs of 20-24 nucleotides in length. As regulators, they play an essential role in biological processes, such as embryonic development (embryogenesis), cell proliferation, cellular differentiation, cell migration, and apoptosis3. Therefore, any change in miRNA expression level may have a link to various diseases like cancer. A number of miRNAs function as both tumor suppressors and as oncogenes (oncomiR)4,5, having been shown to be involved in various cancers including breast cancer6
-8, gastric cancer9
-11, and OSCC12
-14. Contradictory results have been reported about the miRNA expression profiles of OSCC in human societies, and extensive research needs to be conducted before clinical use15,16. MiR-191 is one of the highly expressed and stable miRNAs found in human serum and saliva and is a potential non-invasive biomarker in humans17. MiR-191 is overexpressed in some cancers such as gastric cancer and has been shown to promote cell growth and suppress apoptosis18. MiR-155 plays a prominent role in cancer biology and can be both a suppressor and an oncogene19. As an oncomiR, miR-155 has been recognized as a carcinogen in OSCC20. MiR-494 is also identified as an oncomiR and is a key player in advancement of the cell cycle and proliferation. MiR-494 overexpression decreases PTEN and P27 expression21.
Bevacizumab, sold under the trade name Avastin and approved by the U.S. Food and Drug Administration, is a fully humanized whole monoclonal antibody22 that is used to cure a number of types of cancers, including colorectal cancer, lung cancer23, ovarian cancer, and glioblastoma24. Bevacizumab can act as an angiogenesis inhibitor, cause regulation of miRNA expression, induce cell apoptosis, and prevent OSCC progression25.
This study aims to investigate the change in expression levels of miR-155, miR-191, and miR-494, as diagnostic biomarkers, in the serum of OSCC patients compared to that of healthy participants. To explore the effects of Avastin on apoptosis, the expression levels of miR-155, miR-191, and miR-494, as treatment and diagnostic biomarkers, were reviewed in the HN5 cell line to avoid the unnecessary and exorbitant cost of medical treatments for unresponsive patients.
Go to : 
This case-control study consists of 50 patients who were referred to Tehran University of Medical Sciences, Tehran, Iran, from March 2019 to January 2020. Cases were selected based on physical examination prior to any treatment and were suspected to have OSCC. Another 50 healthy participants were included in this study as the control group after being examined by a doctor and signing the letter of consent (ethical code: IR.TUMS.VCR.REC.1397.792). These people were chosen from the same age groups. Peripheral blood (2 mL) was collected from the 50 OSCC and 50 healthy subjects, and RNA was immediately extracted.
The extraction of total RNA and small RNAs was carried out using RNeasy Mini Kit (Qiagen, Hilden, Germany), according to kit instructions. The quality of the extracted RNA was evaluated by NanoDrop, and the ratio of absorbance at 260 nm and 280 nm was used to assess the RNA concentration. cDNA was produced using a kit from Zistroyesh (Tehran, Iran), real-time reverse transcription polymerase chain reaction (RT-PCR) was performed with the same kit and with a Rotor-Gene (Qiagen, Hilden, Germany). To normalize miRNA expression, we used U6 as a calibrator (housekeeping gene). Temperatures and reaction times were adjusted according to kit instructions.(Table 1) The results were interpreted after each reaction and based on amplification and melting peak curves. Average blood miRNA expression level was calculated using the ΔΔCt method and compared to that of U6 snRNA.
The 2−ΔΔCT method was used to determine the fold change of target miRNA compared to the expressed miRNA in the control group.
The HN5 cell line, under the cellular code C196, was obtained from Pasteur Institute of Iran (Tehran) and transferred to a laboratory at the University of Tehran to be cultured. HN5 cells were incubated and grew in a humidified incubator at 5% CO2 and 37°C. The growth medium, RPMI-1640 (Gibco, Carlsbad, CA, USA), was enriched with phosphate-buffered saline (PBS), 100 U/mL of penicillin, 100 µg/mL of streptomycin, and 10% fetal bovine serum (FBS).
Cell proliferation was assessed using MTT assay. HN5 cells in the exponential phase were placed in 96-well plates. They continued to grow at a concentration of 5×103 cells per well in the RPMI-1640 medium supplemented with 10% PBS. Different concentrations of Avastin (0-650 μM) dissolved in 1% DMSO were added to the HN5 cells and incubated in 5% CO2 at 37°C for 24, 48, and 72 hours. The HN5 cells were then washed with PBS. Next, 200 μL of the culture medium containing 0.5 mg/mL of MTT was added to the cells, which were incubated in 5% CO2 at 37°C for four hours. The cells were lysed after the aforementioned time periods, and purple formazan crystals were measured at 570 nm and 630 nm wavelengths using an ELISA reader. The percentage of cell viability was calculated.
Apoptosis of cells was analyzed using the annexin V/propidium iodide (PI) test. In detail, 1 mL of the cell suspension (equal to 5×105 cells in ml) was poured into six-well plates, and then 2 mL of the growth medium plus 10% FBS were added to the cells. Subsequently, Vanadium Schiff base complex was added to the cells. After the incubation of cells, six-well plates were washed with PBS and centrifuged at room temperature for six minutes. The liquid at the top of the sample was discarded, and 10 μL of annexin was added and incubated on ice in the dark. After 30 minutes, 1 mL of PBS was added to the samples, which were then evaluated by flow cytometry.
Total RNA was extracted using the RNeasy Mini Kit, according to kit instructions. The concentration of extracted RNA was assessed by a NanoDrop, and cDNA was produced using the Zistroyesh Kit. Real-time RT-PCR was performed according to the protocols mentioned in the previous stage.
The sample size was calculated considering marker positivity in both groups, and type I and type II errors were perceived to be 5% and 20%, respectively. Results were analyzed by IBM SPSS Statistics (ver. 20; IBM, Armonk, NY, USA), and the mean and standard deviation were measured. To analyze the differences between the sample and control groups or the link between miRNA expression levels and clinical and pathological features, paired sample t-test was applied. P-values less than 0.05 were statistically significant.
Go to : 
In this research, the statistical population consisted of 50 OSCC patients and 50 healthy participants. These two groups were age-matched. Groups were compared according to age using the t-test, and no significant difference was found; therefore, it was concluded that age would not be an issue in this study.(Table 2) The patient group consisted of 26 males (52.0%) and 24 females (48.0%) individuals, while the healthy group included 28 males (56.0%) and 22 females (44.0%) individuals.
All the real-time RT-PCR reactions were performed with duplicate repetitions. Two vials of cDNA from each sample were made and tested. The results were interpreted according to the melting curve.
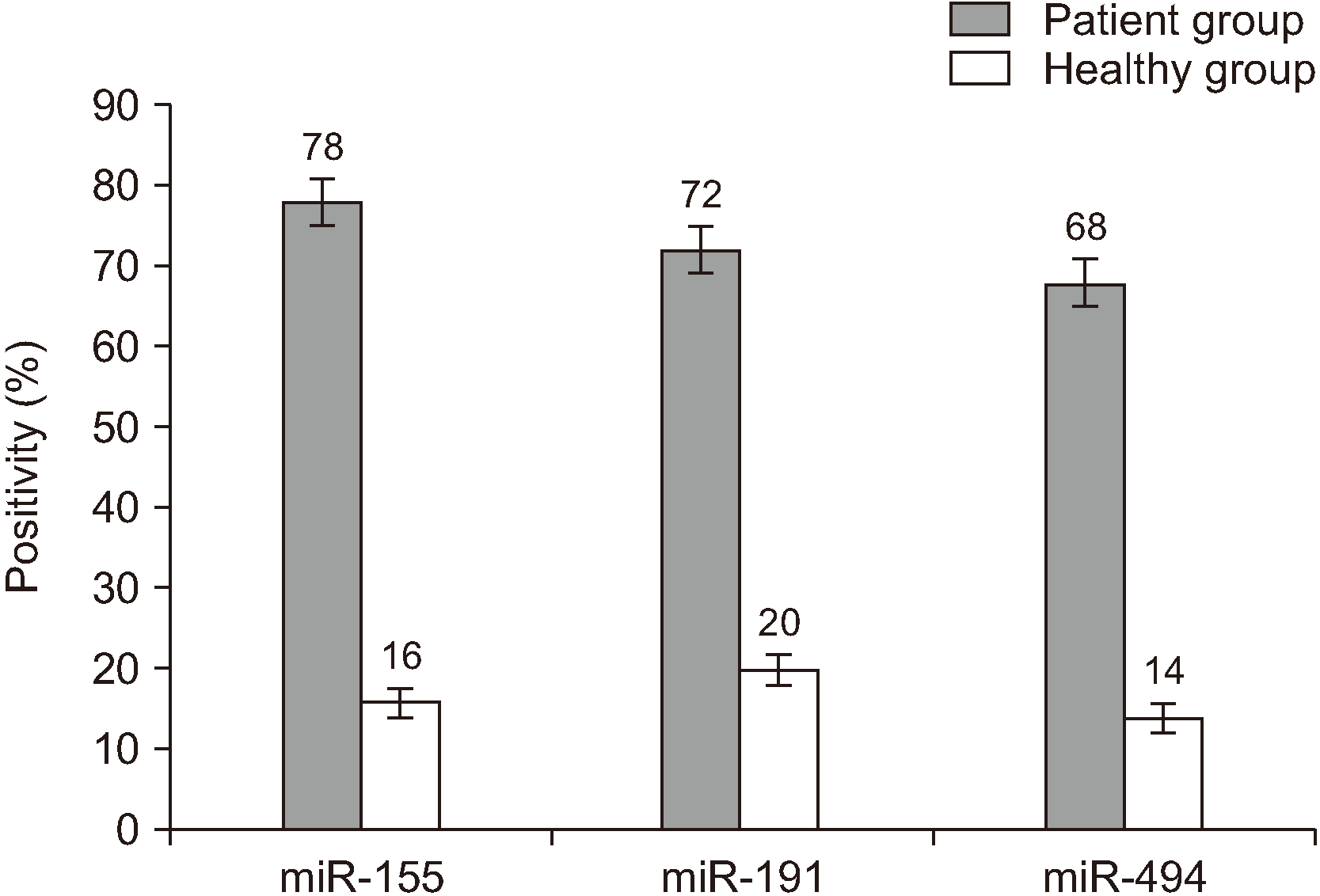
The miR-155, miR-191, and miR-494 biomarkers were positive in 39 (78.0%), 36 (72.0%), and 34 (68.0%) of 50 OSCC-affected participants, respectively, and their numbers in healthy individuals were 8 (16.0%), 10 (20.0%), and 7 (14.0%).(Fig. 1) The statistical comparison of these biomarkers in the sick and healthy groups was conducted by two-sample binomial, which indicated a statistically significant difference between these two groups (P<0.001).
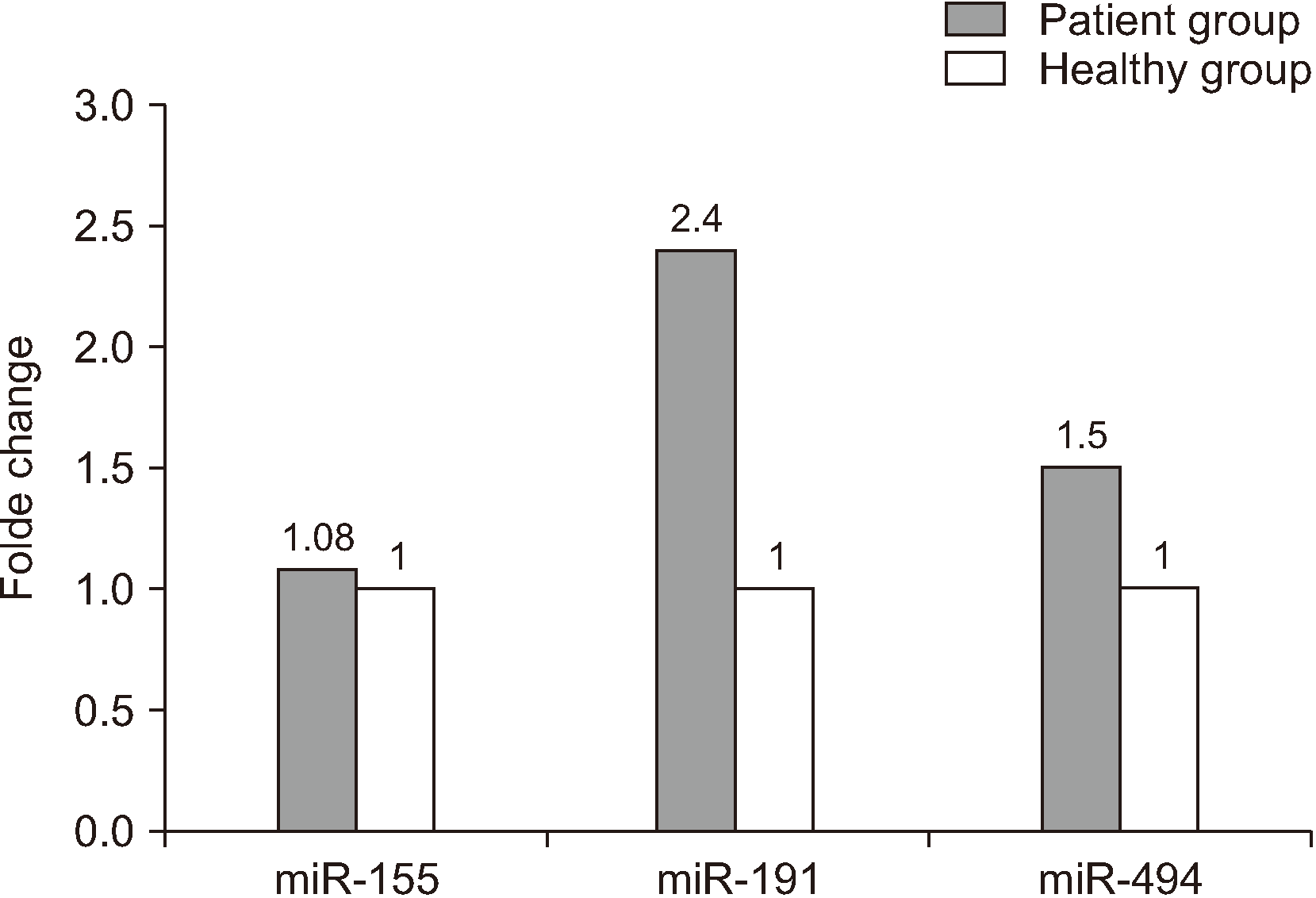
First, cycle threshold (Ct) of each sample (as mentioned before) was determined. The comparative differences between the test and control samples were calculated using the 2–ΔΔCt formula. In peripheral blood, the expression of miR-191, miR-494, and miR-155 biomarkers was on average 2.4, 1.5, and 1.08 times higher, respectively, in sick cases than in healthy individuals.(Fig. 2)
The comparison of cell viability at 0 µM concentration of Avastin at three times showed significant differences (P<0.001). The longer was the elapsed time, the greater was the cell growth. The survival of HN5 cells in the 100 µM concentration of Avastin also differed significantly with time (P=0.02), although that at the 200 µM concentration of Avastin showed no significant difference (P=0.35). The survival of HN5 cells in the 400 µM concentration of Avastin after 48 hours significantly decreased compared to that in the first 24 hours (P=0.001), and a significant reduction was observed in the 600 µM concentration of Avastin after 72 hours compared to the first 24 and 48 hours (P=0.005 and 0.05, respectively).
After incubation of the HN5 cell line in the 0, 100, 200, 400, and 600 µM concentrations of Avastin, apoptosis of HN5 cells was analyzed using flow cytometry with annexin V and PI after 24, 48, and 72 hours. In the 400 µM concentration, the average percentage of apoptotic cells in the control samples was 10% at each time marker, while that in the test samples was 14.8%, 33.4%, and 38.8% at the 24-, 48-, and 72-hour time markers, respectively, which indicated a 1.5-fold, 3.5-fold, and 4-fold increase in apoptosis in the test samples compared to the controls.
The expression of miR-155, miR-191, and miR-494 in HN5 cells were analyzed using real-time PCR after 48 hours incubation with different concentrations (0, 100, 200, 400, and 600 µM) of Avastin. The result indicated the reducing effect of Avastin on the relevant miRNA expression and the positive effect of the medicine on treatment.(Table 3)
Go to : 
There are a number of studies on the role of miRNA and miRNA dysregulation in different diseases, including cancers. MiRNAs are endogenously expressed small non-coding RNAs that play a major role in almost all biological cellular pathways. Thus, they affect many cancer-related cellular processes, such as proliferation, cell cycle control, apoptosis, differentiation, cell migration, and cell metabolism26,27. Many studies have reported that miRNAs can operate as oncogenes or tumor suppressor genes27,28.
Various strategies exist for diagnosing OSCC in later stages and after metastasis, such as sputum cytology and endoscopy. Therefore, there is a compelling reason to find a proper and non-invasive method such as use of tumor biomarkers to enable diagnosis in earlier stages29. The unnatural expression patterns of miRNAs probably have valuable information as important biomarkers for diagnosis, treatment, and prognosis30.
In this study, we showed that the expression of miR-155, miR-191, and miR-494 in the peripheral blood of those with OSCC increased 1.08-fold, 2.4-fold, and 1.5-fold, respectively, so they can be considered as biomarkers with high diagnosis potential in early stages. Furthermore, after 24, 48, and 72 hours of incubation of HN5 cells with Avastin concentrations of 0, 100, 200, 400, and 600 µM, in the 400 µM concentration, the percentage of apoptotic cells in the control samples was 10% on average and that in test samples was 14.8%, 33.4%, (P=0.003), and 38.8% (P=0.02) after 24, 48, and 72 hours, respectively. This finding shows a 1.5-fold, 3.5-fold, and 4-fold increase in apoptosis in the test samples compared to the controls. Meanwhile, following real-time PCR in HN5 cells after 48 hours incubation with Avastin at concentrations of 100, 200, 400, and 600 µM, the levels of overexpressed miR-155, miR-191, and miR-494 decreased, indicating a positive effect of the medicine on treatment. These results suggest that the three miRNAs mentioned could be good markers for diagnosis and follow-up of treatment.
Evidence has revealed changes in expression of miR-155 in carcinogenesis. Overexpression of miR-155 has also been demonstrated in breast cancer6, non-small cell lung cancer (NSCLC)31, gastric cancer9, and osteosarcoma32, which is in agreement with our results. However, in the Zare et al.33 study on gastric cancer, statistically significant downregulation of miR-155-5p (P=0.0018) was reported, which is contradictory to the result obtained here.
MiR-191 dysregulation in different cancers, such as in hepatocellular carcinoma34, breast cancer7, pancreatic cancer35, gastric cancer10, different human cell lines of colorectal cancer36, and OSCC13, has increasingly been detected, which is in agreement with the overexpression of miR-191 in OSCC cases in this study. However, downregulation of miR-191 in follicular adenoma, anaplastic thyroid carcinoma, and papillary thyroid carcinoma37 and downregulation of miR-191-5p in the renal cell carcinoma cell lines ACHN and 786-O38 were in conflict with the results of this study.
MiR-494 expression in breast cancer cell lines MDA-MB-468 and MDA-MB-23139, NSCLC cancer tissues and cell line8, hepatocellular carcinoma40, and colorectal cancer41 increases, which corresponds with our result regarding miR-494 overexpression in OSCC. However, some studies have indicated a connection between miR-494 downregulation and cervical tumors, ovarian tumors42,43, and gastric cancer11, which contradicts our findings.
This study lends support to the results obtained by Zhao et al.44 concerning the anti-tumor effects of Avastin/bevacizumab on the human colorectal cancer HT-29 cell line. In this research, the anti-tumor effects of Avastin was in line with the results achieved in the field of effective and improving effects in treatment of patients affected by different types of tumors such as metastatic renal cell cancer, NSCLC, pancreatic cancer, breast cancer24. Induction of apoptosis and prevention of tumor growth and liver metastasis in advanced gastric cancer are also among the antitumor effects of Avastin45.
In the present investigation, our findings reflect the changes in expression patterns of miR-155, miR-191, and miR-494 in OSCC. Avastin/bevacizumab controls cell proliferation, induces apoptosis, and decreases the viability of cells in the HN5 cancer cell line while also decreasing the expression levels of miR-155, miR-191, and miR-494. Therefore, it seems that the expression levels of the three mRNAs in the blood of those affected by OSCC could serve as diagnostic biomarkers in the early stages of disease.
Go to : 
In treatment with Avastin in chemotherapy, miR-155, miR-191, and miR-494 can be regarded as biomarkers for progression of treatment. Avastin functions as an anti-tumor drug that induces apoptosis in the HN5 cell line and downregulates the expression of the aforementioned biomarkers in OSCC.
Go to : 
Acknowledgements
This study was funded by Tehran University of Medical Sciences (TUMS) (grant No. 97-03-106-39755). Further thanks to the Islamic Azad University, North Tehran branch.
Go to : 
Notes
Authors’ Contributions
A.M. and N.B. conceived the idea, N.B. analyzed the data, and M.B. drafted the article. M.M. and F.M. helped with research procedures. N.E. and A.M. collected the data and performed the research.
Ethics Approval and Consent to Participate
All samples were obtained with patients providing extensively informed consent, and the study was approved by the Tehran University of Medical Sciences ethical committee (ethical code: IR.TUMS.VCR.REC.1397.792).
Go to : 
References
1. Kumar M, Nanavati R, Modi TG, Dobariya C. 2016; Oral cancer: etiology and risk factors: a review. J Cancer Res Ther. 12:458–63.
https://doi.org/10.4103/0973-1482.186696
. DOI: 10.4103/0973-1482.186696. PMID: 27461593.

2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. 2015; Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–86.
https://doi.org/10.1002/ijc.29210
. DOI: 10.1002/ijc.29210. PMID: 25220842.

3. Peng Y, Croce CM. 2016; The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 1:15004.
https://doi.org/10.1038/sigtrans.2015.4
. DOI: 10.1038/sigtrans.2015.4. PMID: 29263891. PMCID: PMC5661652.

4. Nagpal N, Kulshreshtha R. 2014; miR-191: an emerging player in disease biology. Front Genet. 5:99.
https://doi.org/10.3389/fgene.2014.00099
. DOI: 10.3389/fgene.2014.00099. PMID: 24795757. PMCID: PMC4005961.

5. Stahlhut Espinosa CE, Slack FJ. 2006; The role of microRNAs in cancer. Yale J Biol Med. 79:131–40. PMID: 17940623. PMCID: PMC1994807.
6. Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. 2012; Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 7:e47003.
https://doi.org/10.1371/journal.pone.0047003
. DOI: 10.1371/journal.pone.0047003. PMID: 23071695. PMCID: PMC3468565.

7. Zhang X, Wu M, Chong QY, Zhang W, Qian P, Yan H, et al. 2018; Amplification of hsa-miR-191/425 locus promotes breast cancer proliferation and metastasis by targeting DICER1. Carcinogenesis. 39:1506–16.
https://doi.org/10.1093/carcin/bgy102
. DOI: 10.1093/carcin/bgy102. PMID: 30084985.

8. Chen J, Nie S, Hong B, Li C, Xiong T, Shen X, et al. 2017; MicroRNA-494 promotes tumor growth by targeting PTEN in non-small cell lung cancer. Int J Clin Exp Pathol. 10:4441–50.
9. Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, et al. 2018; MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-β receptor 2. Cancer Sci. 109:618–28.
https://doi.org/10.1111/cas.13472
. DOI: 10.1111/cas.13472. PMID: 29247570. PMCID: PMC5834794.

10. Peng WZ, Ma R, Wang F, Yu J, Liu ZB. 2014; Role of miR-191/425 cluster in tumorigenesis and diagnosis of gastric cancer. Int J Mol Sci. 15:4031–48.
https://doi.org/10.3390/ijms15034031
. DOI: 10.3390/ijms15034031. PMID: 24603541. PMCID: PMC3975382.

11. He W, Li Y, Chen X, Lu L, Tang B, Wang Z, et al. 2014; miR-494 acts as an anti-oncogene in gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol. 29:1427–34.
https://doi.org/10.1111/jgh.12558
. DOI: 10.1111/jgh.12558. PMID: 24612089.

12. Shi LJ, Zhang CY, Zhou ZT, Ma JY, Liu Y, Bao ZX, et al. 2015; MicroRNA-155 in oral squamous cell carcinoma: overexpression, localization, and prognostic potential. Head Neck. 37:970–6.
https://doi.org/10.1002/hed.23700
. DOI: 10.1002/hed.23700. PMID: 24692283.

13. Gissi DB, Morandi L, Gabusi A, Tarsitano A, Marchetti C, Cura F, et al. 2018; A noninvasive test for MicroRNA expression in oral squamous cell carcinoma. Int J Mol Sci. 19:1789.
https://doi.org/10.3390/ijms19061789
. DOI: 10.3390/ijms19061789. PMID: 29914173. PMCID: PMC6032413.

14. Libório-Kimura TN, Jung HM, Chan EK. 2015; miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 51:151–7.
https://doi.org/10.1016/j.oraloncology.2014.11.019
. DOI: 10.1016/j.oraloncology.2014.11.019. PMID: 25500095.

15. Zahra A, Rubab I, Malik S, Khan A, Khan MJ, Fatmi MQ. 2018; Meta-analysis of miRNAs and their involvement as biomarkers in oral cancers. Biomed Res Int. 2018:8439820.
https://doi.org/10.1155/2018/8439820
. DOI: 10.1155/2018/8439820. PMID: 29516011. PMCID: PMC5817319.

16. Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo Muzio L. 2018; Predictive prognostic value of tissue-based microRNA expression in oral squamous cell carcinoma: a systematic review and meta-analysis. J Dent Res. 97:759–66.
https://doi.org/10.1177/0022034518762090
. DOI: 10.1177/0022034518762090. PMID: 29533734.

17. Patel RS, Jakymiw A, Yao B, Pauley BA, Carcamo WC, Katz J, et al. 2011; High resolution of microRNA signatures in human whole saliva. Arch Oral Biol. 56:1506–13.
https://doi.org/10.1016/j.archoralbio.2011.05.015
. DOI: 10.1016/j.archoralbio.2011.05.015. PMID: 21704302. PMCID: PMC3189266.

18. Shi X, Su S, Long J, Mei B, Chen Y. 2011; MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim Biophys Sin (Shanghai). 43:849–56.
https://doi.org/10.1093/abbs/gmr084
. DOI: 10.1093/abbs/gmr084. PMID: 21947487.

19. Bedewy AML, Elmaghraby SM, Shehata AA, Kandil NS. 2017; Prognostic value of miRNA-155 expression in B-cell non-hodgkin lymphoma. Turk J Haematol. 34:207–12.
https://doi.org/10.4274/tjh.2016.0286
. DOI: 10.4274/tjh.2016.0286. PMID: 28148469. PMCID: PMC5544039.

20. Iorio MV, Croce CM. 2012; microRNA involvement in human cancer. Carcinogenesis. 33:1126–33.
https://doi.org/10.1093/carcin/bgs140
. DOI: 10.1093/carcin/bgs140. PMID: 22491715. PMCID: PMC3514864.

21. Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, et al. 2018; The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 9:4.
https://doi.org/10.1038/s41419-017-0076-6
. DOI: 10.1038/s41419-017-0076-6. PMID: 29305580. PMCID: PMC5849044.

22. Shih T, Lindley C. 2006; Bevacizumab: an angiogenesis inhibitor for the treatment of solid malignancies. Clin Ther. 28:1779–802.
https://doi.org/10.1016/j.clinthera.2006.11.015
. DOI: 10.1016/j.clinthera.2006.11.015. PMID: 17212999.

23. Pang W, Su J, Wang Y, Feng H, Dai X, Yuan Y, et al. 2015; Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci. 106:1362–9.
https://doi.org/10.1111/cas.12747
. DOI: 10.1111/cas.12747. PMID: 26195069. PMCID: PMC4638007.

24. de Gramont A, Van Cutsem E. 2005; Investigating the potential of bevacizumab in other indications: metastatic renal cell, non-small cell lung, pancreatic and breast cancer. Oncology. 69 Suppl 3:46–56.
https://doi.org/10.1159/000088483
. DOI: 10.1159/000088483. PMID: 16301835.

25. Yoshida H, Yoshimura H, Matsuda S, Ryoke T, Kiyoshima T, Kobayashi M, et al. 2018; Effects of peritumoral bevacizumab injection against oral squamous cell carcinoma in a nude mouse xenograft model: a preliminary study. Oncol Lett. 15:8627–34.
https://doi.org/10.3892/ol.2018.8399
. DOI: 10.3892/ol.2018.8399. PMID: 29805597. PMCID: PMC5950523.

26. Zhao XM, Liu KQ, Zhu G, He F, Duval B, Richer JM, et al. 2015; Identifying cancer-related microRNAs based on gene expression data. Bioinformatics. 31:1226–34.
https://doi.org/10.1093/bioinformatics/btu811
. DOI: 10.1093/bioinformatics/btu811. PMID: 25505085.

27. Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. 2016; microRNA therapeutics in cancer - an emerging concept. EBioMedicine. 12:34–42.
https://doi.org/10.1016/j.ebiom.2016.09.017
. DOI: 10.1016/j.ebiom.2016.09.017. PMID: 27720213. PMCID: PMC5078622.

28. Hammond SM. 2006; MicroRNAs as oncogenes. Curr Opin Genet Dev. 16:4–9.
https://doi.org/10.1016/j.gde.2005.12.005
. DOI: 10.1016/j.gde.2005.12.005. PMID: 16361094.

29. Hutvágner G, Zamore PD. 2002; A microRNA in a multiple-turnover RNAi enzyme complex. Science. 297:2056–60.
https://doi.org/10.1126/science.1073827
. DOI: 10.1126/science.1073827. PMID: 12154197.

30. Kroh EM, Parkin RK, Mitchell PS, Tewari M. 2010; Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 50:298–301.
https://doi.org/10.1016/j.ymeth.2010.01.032
. DOI: 10.1016/j.ymeth.2010.01.032. PMID: 20146939. PMCID: PMC4186708.

31. Xue X, Liu Y, Wang Y, Meng M, Wang K, Zang X, et al. 2016; MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 7:84508–19.
https://doi.org/10.18632/oncotarget.13022
. DOI: 10.18632/oncotarget.13022. PMID: 27811366. PMCID: PMC5356677.

32. Lu S, Liao QS, Tang L. 2018; MiR-155 affects osteosarcoma cell proliferation and invasion through regulating NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 22:7633–9.
https://doi.org/10.26355/eurrev_201811_16380
. DOI: 10.26355/eurrev_201811_16380. PMID: 30536304.

33. Zare A, Alipoor B, Omrani MD, Zali MR, Malekpour Alamdari N, Ghaedi H. 2019; Decreased miR-155-5p, miR-15a, and miR-186 expression in gastric cancer is associated with advanced tumor grade and metastasis. Iran Biomed J. 23:338–43.
https://doi.org/10.29252/.23.5.338
. DOI: 10.29252/ibj.23.5.5. PMID: 31103022. PMCID: PMC6661124.

34. Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, et al. 2010; hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 70:8077–87.
https://doi.org/10.1158/0008-5472.CAN-10-1313
. DOI: 10.1158/0008-5472.CAN-10-1313. PMID: 20924108.

35. Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Lu J, et al. 2014; MicroRNA-191 promotes pancreatic cancer progression by targeting USP10. Tumour Biol. 35:12157–63.
https://doi.org/10.1007/s13277-014-2521-9
. DOI: 10.1007/s13277-014-2521-9. PMID: 25168367.

36. Zhang XF, Li KK, Gao L, Li SZ, Chen K, Zhang JB, et al. 2015; miR-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPβ. Oncotarget. 6:4144–58.
https://doi.org/10.18632/oncotarget.2864
. DOI: 10.18632/oncotarget.2864. PMID: 25784653. PMCID: PMC4414178.

37. Colamaio M, Borbone E, Russo L, Bianco M, Federico A, Califano D, et al. 2011; miR-191 down-regulation plays a role in thyroid follicular tumors through CDK6 targeting. J Clin Endocrinol Metab. 96:E1915–24.
https://doi.org/10.1210/jc.2011-0408
. DOI: 10.1210/jc.2011-0408. PMID: 21956418.

38. Chen P, Pan X, Zhao L, Jin L, Lin C, Quan J, et al. 2018; MicroRNA-191-5p exerts a tumor suppressive role in renal cell carcinoma. Exp Ther Med. 15:1686–93.
https://doi.org/10.3892/etm.2017.5581
. DOI: 10.3892/etm.2017.5581. PMID: 29434754. PMCID: PMC5774385.

39. Macedo T, Silva-Oliveira RJ, Silva VAO, Vidal DO, Evangelista AF, Marques MMC. 2017; Overexpression of mir-183 and mir-494 promotes proliferation and migration in human breast cancer cell lines. Oncol Lett. 14:1054–60.
https://doi.org/10.3892/ol.2017.6265
. DOI: 10.3892/ol.2017.6265. PMID: 28693273. PMCID: PMC5494613.

40. Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, et al. 2015; miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 34:1003–10.
https://doi.org/10.3892/or.2015.4030
. DOI: 10.3892/or.2015.4030. PMID: 26045065.

41. Zhang Y, Guo L, Li Y, Feng GH, Teng F, Li W, et al. 2018; MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer. 17:1.
https://doi.org/10.1186/s12943-017-0753-1
. DOI: 10.1186/s12943-017-0753-1. PMID: 29304823. PMCID: PMC5755155.

42. Yang YK, Xi WY, Xi RX, Li JY, Li Q, Gao YE. 2015; MicroRNA-494 promotes cervical cancer proliferation through the regulation of PTEN. Oncol Rep. 33:2393–401.
https://doi.org/10.3892/or.2015.3821
. DOI: 10.3892/or.2015.3821. PMID: 25738254.

43. Yuan J, Wang K, Xi M. 2016; MiR-494 inhibits epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monit. 22:617–24.
https://doi.org/10.12659/msm.897288
. DOI: 10.12659/MSM.897288. PMID: 26908019. PMCID: PMC4768945.

44. Zhao Z, Xia G, Li N, Su R, Chen X, Zhong L. 2018; Autophagy inhibition promotes bevacizumab-induced apoptosis and proliferation inhibition in colorectal cancer cells. J Cancer. 9:3407–16.
https://doi.org/10.7150/jca.24201
. DOI: 10.7150/jca.24201. PMID: 30271503. PMCID: PMC6160673.

45. Kang YK, Kang WK, Shin DB, Chen J, Xiong J, Wang J, et al. 2009; Capecitabine/cisplatin versus 5-fluorouracil/cisplatin as first-line therapy in patients with advanced gastric cancer: a randomised phase III noninferiority trial. Ann Oncol. 20:666–73.
https://doi.org/10.1093/annonc/mdn717
. DOI: 10.1093/annonc/mdn717. PMID: 19153121.

Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download