I. Introduction
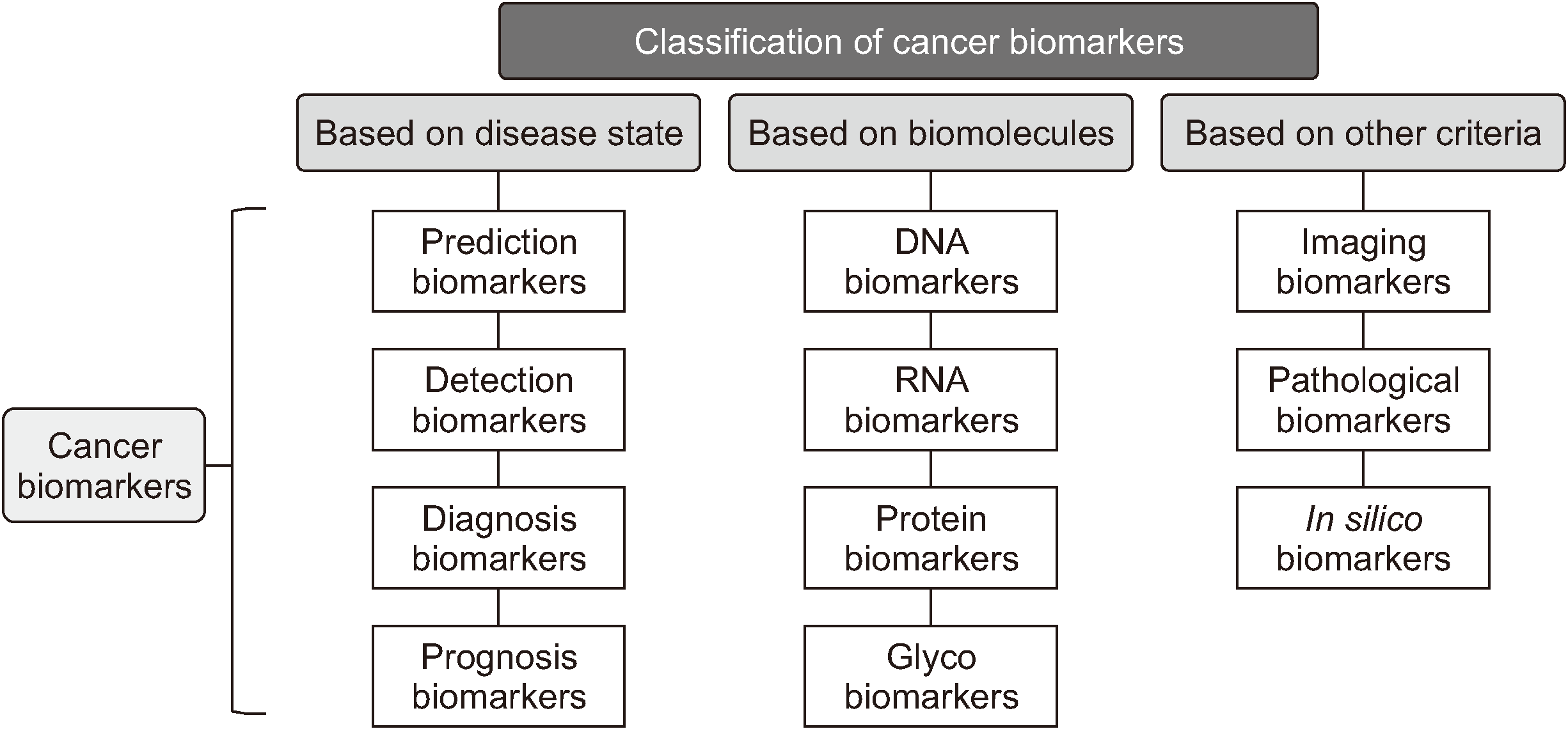 | Fig. 1Classification of biomarkers. Adapted from the article of Mishra and Verma
1
(Cancers [Basel] 2010;2:190-208) in accordance with the Creative Commons Attribution 3.0 Unported (CC BY 3.0) license. |
II. Salivary Biomarkers
Table 1
Table 2
| Disease | Type of saliva | Proteomic approach | Proteins identified | References |
|---|---|---|---|---|
| Oral squamous cell carcinoma | USWS | Mass spectrometry (MS) and western blotting | Increased abundance of myosin and actin. | de Jong et al. 21 |
| USWS | Using shotgun proteomics approach (RP-HPLC, CP-LC with TOF and immunoassay) | Detection of 52 protein that presented in diseased samples but absent in healthy samples | Hu et al. 25 | |
| USWS | Using ultraperformance liquid chromatography-mass spectrometry (UPLC-MS) with hydrophilic interaction chromatography mode |
↑Level of choline, betaine and pipecolinic acid ↑Level of L-carnitine |
Wang et al.22 | |
| Oral leukoplakia | USWS | Two-dimensional gel electrophoresis, mass spectrometry, immunohistochemistry | 22 spots very abundant among them apolipoprotein A1, alpha-amylase, cystatins, keratin 10, lysozyme precursor, and CK10 were relevant to the study. | Camisasca et al. 23 |
| Proliferative verrucous leukoplakia | USWS | Mass spectrometry | Angiotensinogen (AGT) and dipeptidyl peptidase 1 (DPP1) | Flores et al. 24 |
| Premalignant lesions | USWS | Western blotting, mass spectrometry | Salivary actin and myosin | de Jong et al. 21 |
Table 3
| Candidate biomarkers | Techniques | Clinical significance | References |
|---|---|---|---|
| Interleukin (IL)-6, IL-8, IL-1α, IL-1β, TNF-α, tissue polypeptide antigen (TPA), Cyfra 21-1, cancer antigen 125 (CA 125), telomerase, Mac-2 binding protein (M2BP) | ELISA |
Proinflammatory and proangiogenic cytokines found to be indicators of carcinogenic transformation from oral precancerous lesions to oral cancer. Cyfra 21-1, CA 125, and TPA markers attend in telomerase activity in tumor cells and are responsible for the maintenance of telomere length. M2BP helps in the detection of OSCC. |
Katakura et al.
28
Duffy et al. 29 Zhong et al. 30 Krishna Prasad et al. 31 |
| CD44, CD59, Profilin, MRP14 | Immunoblot |
CD44 and CD59 are the very high sensitive cancer and benign diseases differentiate markers. MRP14 is a calcium-binding protein with a sensitivity of 90% and a specificity of 83% in cancer detection. |
Hu et al.
25
Franzmann et al. 32 |
| Glutathione | HPLC | Epidemiological marker for chemoprevention identifies the risk of development of OSCC. | Almadori et al. 26 |
| Mac-2 binding protein (M2BP), Squamous cell carcinoma antigen 2, involucrin, calcyclin, cathepsin-G, azurocidin, transaldolase, carbonic anhydrase I, calgizzarin, myeloblastin, vitamin D-binding protein | ELISA, shotgun proteomics | M2BP is for detection of OSCC this biomarker gives a sensitivity of 90% and a specificity of 83%, and all of them serve as a clinical tool for the noninvasive diagnosis of OSCC. | Hu et al. 25 |
| Immunoglobulin heavy chain constant region gamma (IgG), S100 calciumbinding protein, cofilin-1, transferrin, fibrin | LC/MS | IgG known to be an inhibitor of apoptosis, S100A2, an 11.4 kDa protein that is a prognostic biomarker for OSCC, cofilin proteins are involved in cancer progression, metastasis, and angiogenesis. Transferrin levels in saliva are associated with the size and stage of cancer. Fibrin in OSCC is involved in several carcinogenic processes. |
Jou et al.
27
Kumar et al. 33 |
| α-1-antitrypsin (AAT) | 2DE | AAT is useful for the prediction and determining the aggression of OSCC. | Righini et al. 34 |
| Secretory leukocyte peptidase inhibitor (SLPI), cystatin A, keratin 36, thioredoxin, haptoglobin (HAP), salivary zinc finger, protein 510 peptide, a-amylase, and albumin | MS-based proteomics | SLPI, cystatin A, keratin 36 are potentially involved in the preventive treatment of OSCC. Thioredoxin mRNA levels are elevated in oral cancers and in other cancers as well. Salivary zinc finger, protein 510 peptide, a-amylase, and albumin are useful in the early detection of OSCC. |
Reddy et al.
35
Al Kawas et al. 36 |
Table 4
| Candidate biomarkers | Techniques | Clinical significance | References |
|---|---|---|---|
| Interleukin (IL)-1 , IL-8 | ELISA | Angiogenesis, cell adhesion, chemotaxis, immune response, replication, signal transduction, proliferation, inflammation, and apoptosis |
Li et al.
37
Elashoff et al. 38 |
| Dual specificity phosphatase 1 (DUSP1) | Quantitative PCR (qPCR) and microarrays followed by qPCR | Oxidative stress, protein modification, signal transduction | Li et al. 37 |
| H3 histone family 3A (H3F3A) | qPCR and microarrays followed by qPCR | DNA binding activity | Li et al. 37 |
| Long noncoding HOTAIR | qPCR and microarrays followed by qPCR | Expression of HOTAIR is associated with p53 gene and causes DNA damage | Tang et al. 39 |
| miR-125a, miR-200a, miR-31 | qPCR and microarrays followed by qPCR | Posttranscriptional regulation by RNA silencing complex, cellular growth, and elevated levels in proliferation in OSCC |
Liu et al.
40
Park et al. 41 |
Table 5
| Candidate biomarkers | Techniques | Clinical significance | References |
|---|---|---|---|
| Cadaverine, alanine, serine, glutamine, piperidine, taurine piperidine, choline, pyrroline hydroxycarboxylic acid, beta-alanine, alpha-aminobutyric acid betaine, tyrosine, leucine+isoleucine, histidine, tryptophan, glutamic acid, threonine, carnitine, pipercolic acid, lactic acid, phenylalanine and valine | Capillary electrophoresis time-of-flight mass spectrometry (CE-TOF-MS) and HPLC with quadrupole/TOF MS | Facilitates the clinical detection of OSCC and improves its diagnosis and prognosis. They have a high level of predictive value and serves as a stratification tool. |
Wei et al.
42
Sugimoto et al. 43 |
| Hypoxanthine, guanine, guanosine, trimethylamine N-oxide, spermidine, pipecolate, methionine | CE-TOF-MS | Discrimination of controls from OSCC patients and all of these metabolites have elevated levels in saliva, and hence can be used in noninvasive oral cancer screening. | Ishikawa et al. 44 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download