INTRODUCTION
MATERIALS AND METHODS
Sources of KPC-producing Enterobacteriaceae isolates
Bacterial isolates and antimicrobial susceptibility testing
Detection of resistance genes
Multilocus sequence typing (MLST)
Bacterial conjugation
Curing test
Whole-genome sequencing (WGS)
GenBank accession numbers
RESULTS
Antimicrobial susceptibilities and molecular typing
Table 1
| Isolate I.D | Specimen | Date | MLST | blaKPC | Plasmid | Strain-susceptible antimicrobials† | Carbapenemase differentiation test | OMP loss | Curing test | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||||||||
| ST | Subtype | Bracketed by | pKPC | Replicon | MIC (mg/L) | Zone diameter (mm) | mCIM | eCIM | ||||||||||||
|
|
|
|||||||||||||||||||
| AMK | GEN | CIP | TIG | CST | TMP/SMX | AMK | GEN | CIP | ||||||||||||
| CPKp1825 | Urine | 27-Sep | 307 | blaKPC-2 | ΔTn4401a | pKPBHS25-2 | IncX3 | 16 | 0.25 | 22 | + | - | ompK35, ompK36 | - | ||||||
|
|
||||||||||||||||||||
| CCPKp1825 | blaKPC-2 | ≤ 2 | 0.25 | 22 | ||||||||||||||||
|
|
||||||||||||||||||||
| CPEa1826 | Rectal | 21-Oct | - | blaKPC-2 | ΔTn4401a | pKPBHS26-2 | IncX3 | ≤ 2 | ≤ 1 | ≤ 0.25 | 0.38 | 0.25 | ≤ 20 | 23 | 22 | 30 | + | - | ompK36 | - |
|
|
||||||||||||||||||||
| CCPEa1826 | blaKPC-2 | ≤ 2 | ≤ 1 | ≤ 0.25 | 0.38 | 0.25 | ≤ 20 | 25 | 22 | 30 | ||||||||||
|
|
||||||||||||||||||||
| CPEc1827 | Rectal | 23-Oct | 720 | blaKPC-2 | ΔTn4401a | pKPBHS27-1 | IncX3 | ≤ 2 | ≤ 1 | ≤ 0.25 | 0.125 | 0.25 | ≤ 20 | 20 | 22 | 30 | + | - | ompK35 | - |
|
|
||||||||||||||||||||
| CCPEc1827 | blaKPC-2 | ≤ 2 | ≤ 1 | ≤ 0.25 | 0.75 | 0.25 | ≤ 20 | 23 | 22 | 30 | ||||||||||
* Breakpoints were applied according to the CLSI guidelines [14];
† Disk diffusion test results were interpreted according to the CLSI guidelines [14].
Abbreviations: AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; TIG, tigecycline; CST, colistin; TMP/SMX, trimethoprim-sulfamethoxazole; OMP, outer membrane protein; mCIM, modified carbapenem inactivation method; eCIM, EDTA-modified carbapenem inactivation method; CLSI, Clinical and Laboratory Standards Institute.
Table 2
Sequencing and annotation of CPKp1825
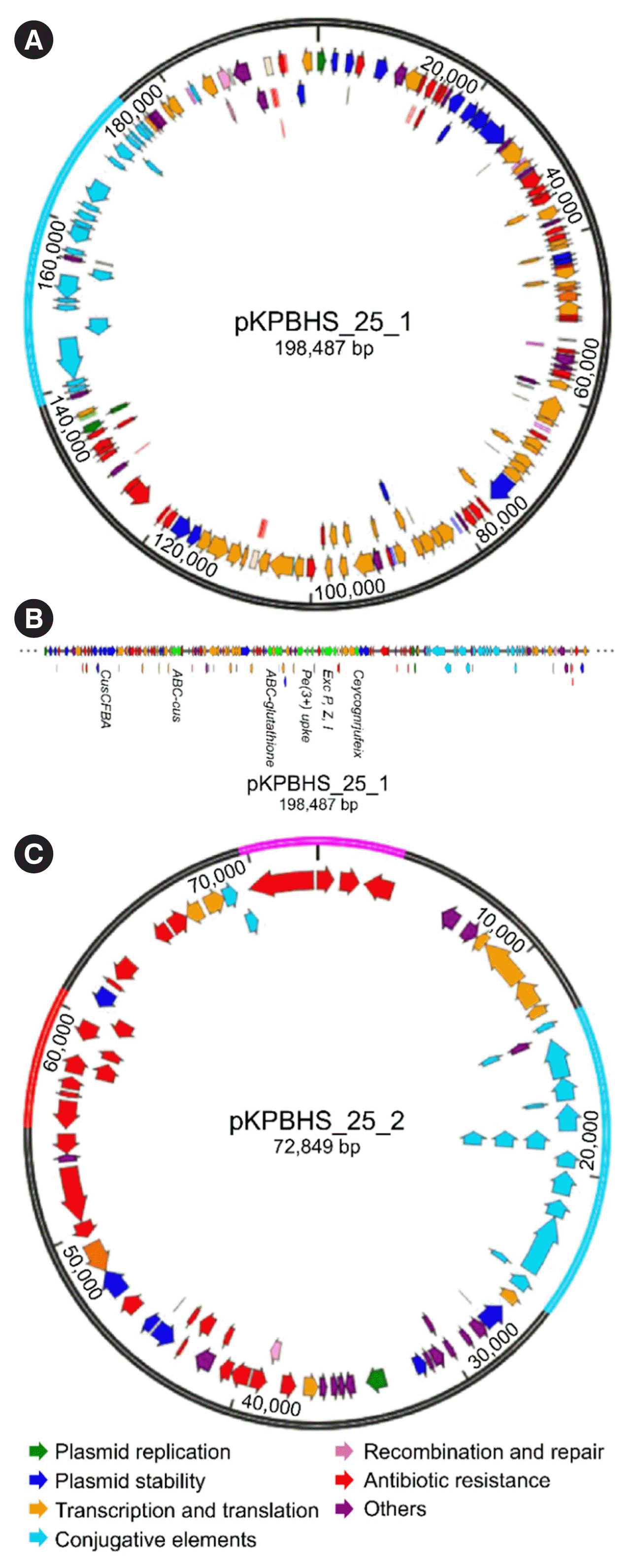 | Fig. 1Genetic organization of plasmids associated with blaKPC. (A) Circular map of pKPBHS_25_1 with two replication origins for the IncFIB (K) and IncFII (K) groups, as well as a copper-transporting efflux system (CusCFBA) and five putative virulence clusters. (B) Variant pKPBHS_25_1 plasmids identified in ST307. (C) Circular map of pECBHS_25_2 containing ΔTn4401a with blaCTX-M-15, blaKPC-2, blaSHV-182, blaTEM-1B, and conjugative elements. Each arrow indicates plasmid scaffold genes and their direction of transcription. The locus Tra is indicated by sky blue arrows, indicating the tra genes (e.g. traG, G; traF, F; traO, O). Related genes (tnpA, tnpR, and tnpM), resistance genes, and insertion sequences are indicated by red arrows. Other genes are indicated by colored arrows as follows: orange, transcription, and translation genes; blue, CusCFBA; green, clusters encoding putative virulence determinants. In the plasmid circle, the Tn3 transposon is indicated in red, ΔTn4401a with blaKPC-2 in pink and conjugative elements in sky blue. |
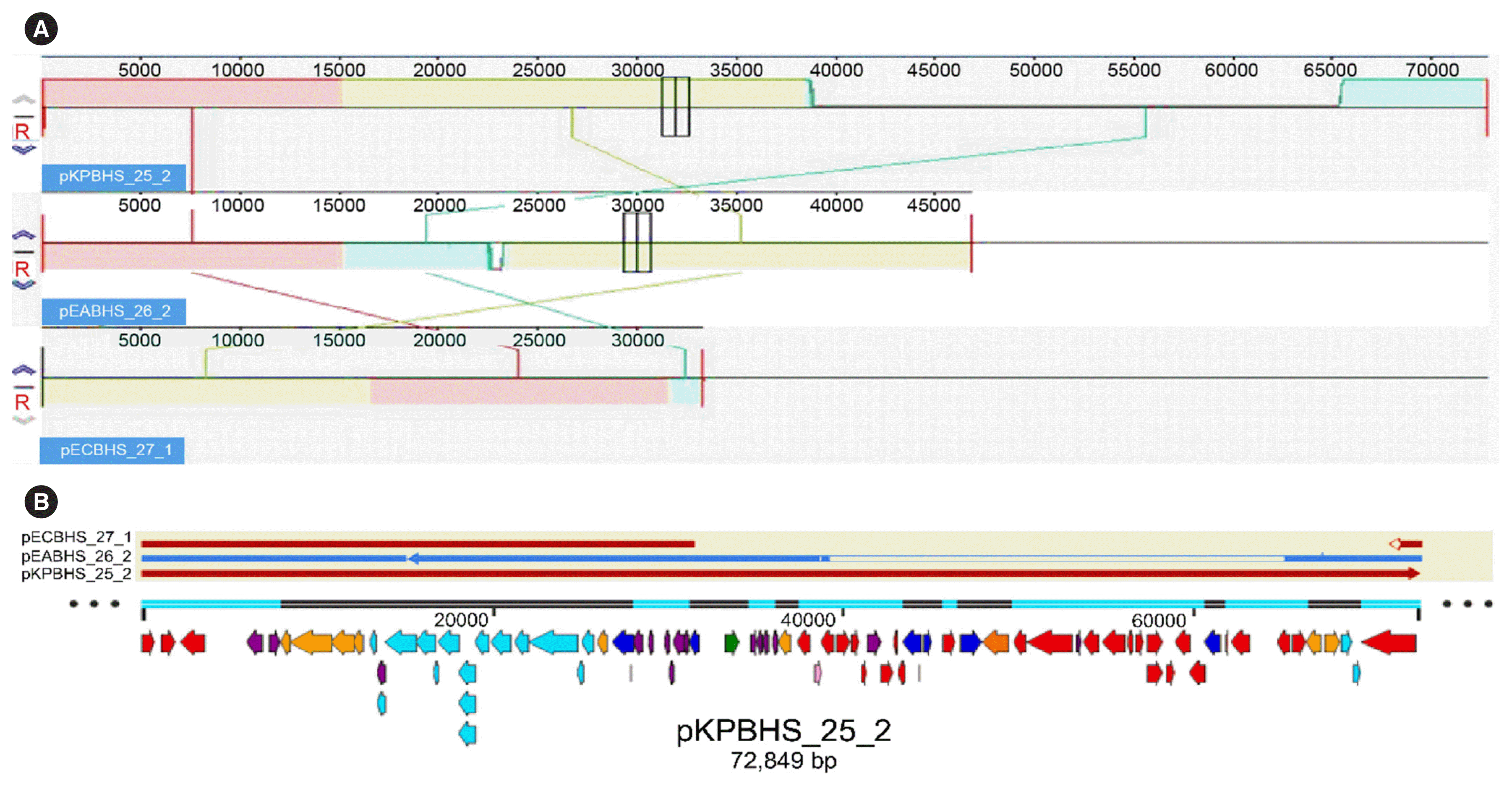 | Fig. 2Linear comparison of the genetic surroundings of blaKPC-2. (A) One fragment unit shows pink, yellow green, and green on the contig. Gene contents were similar along the synteny on the contig. The fragment was well preserved, but inversion existed. Most genes were well preserved. (B) pKPBHS_25_2, pEABHS_26_2, and pECBHS_27_1 are denoted by arrows and colored based on gene-functional classification. Other genes are indicated by colored arrows as follows: green, plasmid replication; blue, plasmid stability; orange, transcription and translation; sky blue, conjugative elements; pink, recombination and repair; red, antimicrobial resistance; purple, other genes. |
Sequencing and annotation of CPEa1826
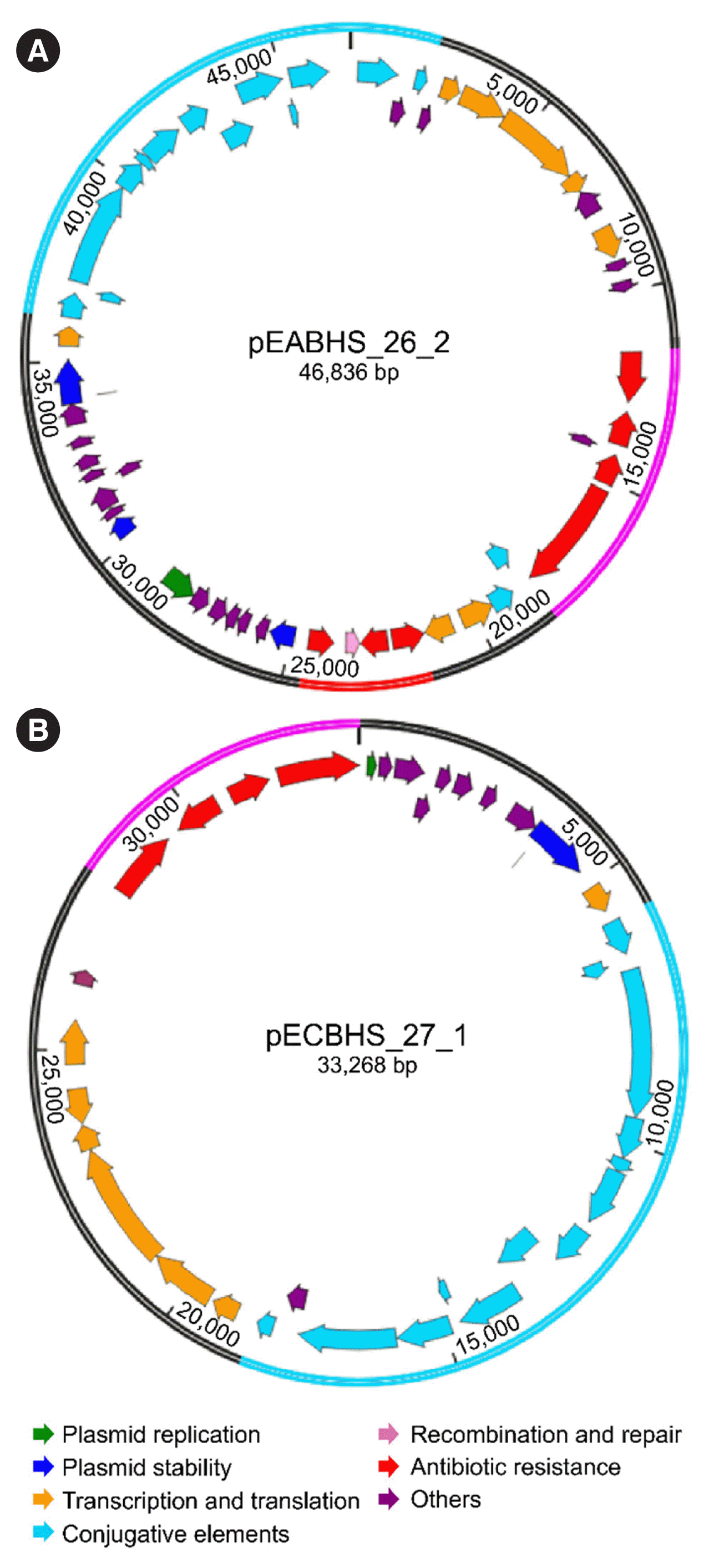 | Fig. 3Genetic organization of plasmids pEABHS_26_2 and pECBHS_27_1. (A) Circular map of pEABHS_26_2 containing ΔTn4401a harboring blaKPC-2 and blaSHV-182 genes. (B) Circular map of pECBHS_27_1 containing ΔTn4401a harboring blaKPC-2 and conjugal transfer genes. In the plasmid circle, the Tn3 transposon is indicated in red, ΔTn4401a with blaKPC-2 in pink and conjugative elements in sky blue. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download