1. Vill K, Kölbel H, Schwartz O, Blaschek A, Olgemöller B, Harms E, et al. One year of newborn screening for SMA–results of a German pilot project. J Neuromuscul Dis. 2019; 6:503–15.


3. Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012; 20:27–32.

4. Monaghan KG, Feldman GL, Palomaki GE, Spector EB. Ashkenazi Jewish Reproductive Screening Working Group, Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. Technical standards and guidelines for reproductive screening in the Ashkenazi Jewish population. Genet Med. 2008; 10:57–72.


5. Grody WW, Thompson BH, Gregg AR, Bean LH, Monaghan KG, Schneider A, et al. ACMG position statement on prenatal/preconception expanded carrier screening. Genet Med. 2013; 15:482–3.


6. Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995; 80:155–65.

7. Bürglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, et al. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996; 32:479–82.

8. Chan V, Yip B, Yam I, Au P, Lin CK, Wong V, et al. Carrier incidence for spinal muscular atrophy in southern Chinese. J Neurol. 2004; 251:1089–93.


9. Su YN, Hung CC, Lin SY, Chen FY, Chern JP, Tsai C, et al. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005–2009: a prospective population-based cohort study. PLoS One. 2011; 6:e17067.

10. Wang KC, Chang CC, Chang YF, Wang SH, Chiang CK, Tsai CP. Evaluation and characterization of a high-resolution melting analysis kit for rapid carrier-screening test of spinal muscular atrophy. J Neurogenet. 2015; 29:113–6.

11. Lundin KE, H⊘jland T, Hansen BR, Persson R, Bramsen JB, Kjems J, et al. Biological activity and biotechnological aspects of locked nucleic acids. Adv Genet. 2013; 82:47–107.


12. Ishige T, Itoga S, Matsushita K. Locked nucleic acid technology for highly sensitive detection of somatic mutations in cancer. Adv Clin Chem. 2018; 83:53–72.


13. Edwards JG, Feldman G, Goldberg J, Gregg AR, Norton ME, Rose NC, et al. Expanded carrier screening in reproductive medicine-points to consider: a joint statement of the American College of Medical Genetics and Genomics, American College of Obstetricians and Gynecologists, National Society of Genetic Counselors, Perinatal Quality Foundation, and Society for Maternal-Fetal Medicine. Obstet Gynecol. 2015; 125:653–62.

14. American College of Obstetricians and Gynecologists. Committee opinion No. 690 summary: carrier screening in the age of genomic medicine. Obstet Gynecol. 2017; 129:e35–e40.
16. Vidal-Folch N, Gavrilov D, Raymond K, Rinaldo P, Tortorelli S, Matern D, et al. Multiplex droplet digital PCR method applicable to newborn screening, carrier status, and assessment of spinal muscular atrophy. Clin Chem. 2018; 64:1753–61.


17. Feng Y, Ge X, Meng L, Scull J, Li J, Tian X, et al. The next generation of population-based spinal muscular atrophy carrier screening: comprehensive pan-ethnic SMN1 copy-number and sequence variant analysis by massively parallel sequencing. Genet Med. 2017; 19:936–44.


18. Mishra S, Lee Y, Park JW. Direct quantification of trace amounts of a chronic myeloid leukemia biomarker using locked nucleic acid capture probes. Anal Chem. 2018; 90:12824–31.


19. Ishige T, Satoh M, Itoga S, Nishimura M, Matsushita K, Nomura F. High-throughput genotyping of GC (vitamin D-binding protein) by melting analysis with locked nucleic acid-incorporating dual hybridization probe for improving mismatch discrimination. Clin Chim Acta. 2018; 487:126–32.

20. Taylor JL, Lee FK, Yazdanpanah GK, Staropoli JF, Liu M, Carulli JP, et al. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin Chem. 2015; 61:412–9.


21. Chien YH, Chiang SC, Weng WC, Lee NC, Lin CJ, Hsieh WS, et al. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J Pediatr. 2017; 190:124–9.e1.


22. Gong B, Zhang L, Hou YP, Hu HY, Li HC, Tan MY, et al. Carrier screening for spinal muscular atrophy in 4719 pregnant women in Shanghai region [Article in Chinese]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013; 30:670–2.

24. Mailman MD, Hemingway T, Darsey RL, Glasure CE, Huang Y, Chadwick RB, et al. Hybrids monosomal for human chromosome 5 reveal the presence of a spinal muscular atrophy (SMA) carrier with two SMN1 copies on one chromosome. Hum Genet. 2001; 108:109–15.

25. Wei X, Tan H, Yang P, Zhang R, Tan B, Zhang Y, et al. Notable carrier risks for individuals having two copies of
SMN1 in spinal muscular atrophy families with 2-copy alleles: estimation based on Chinese meta-analysis data. J Genet Couns. 2017; 26:72–8.

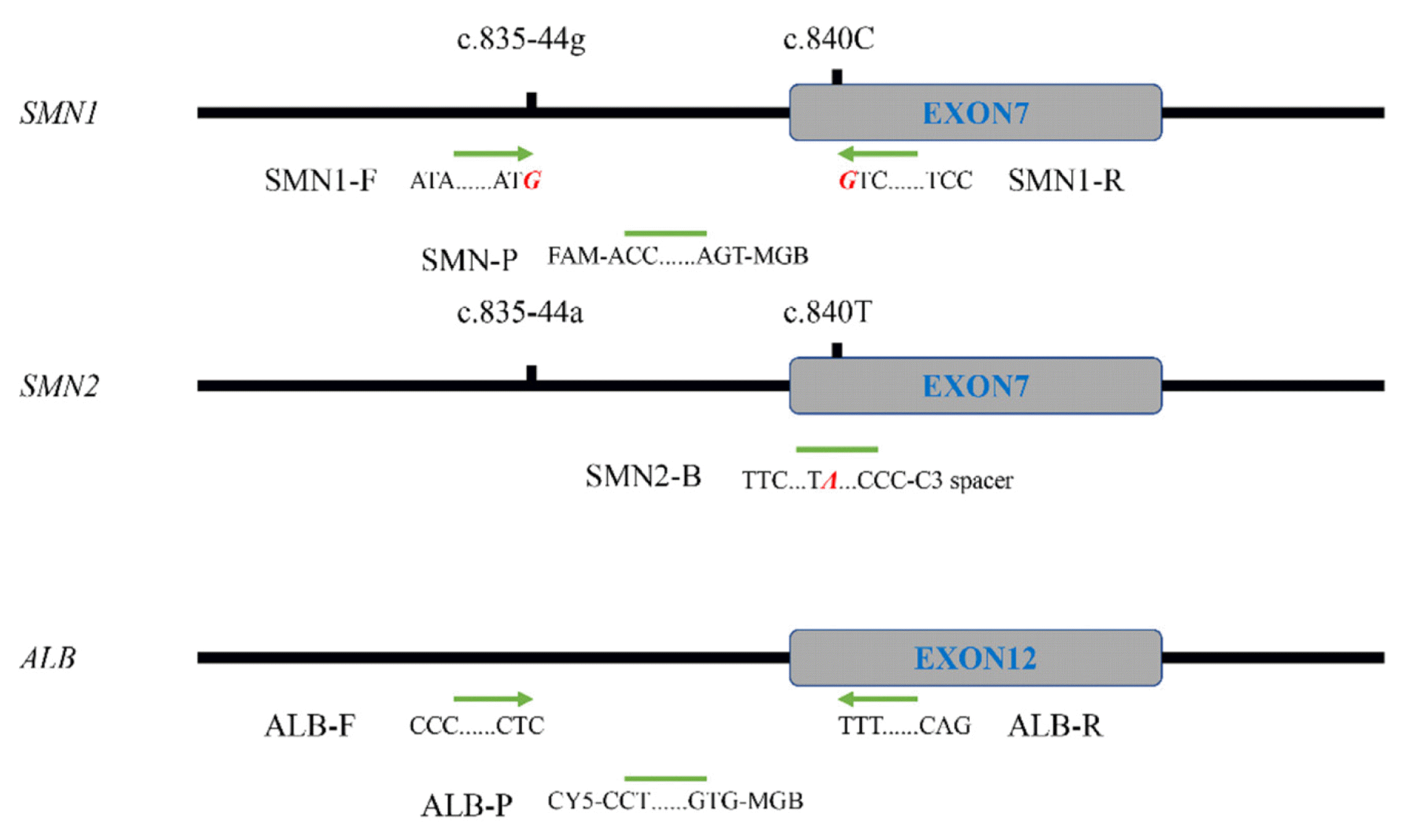
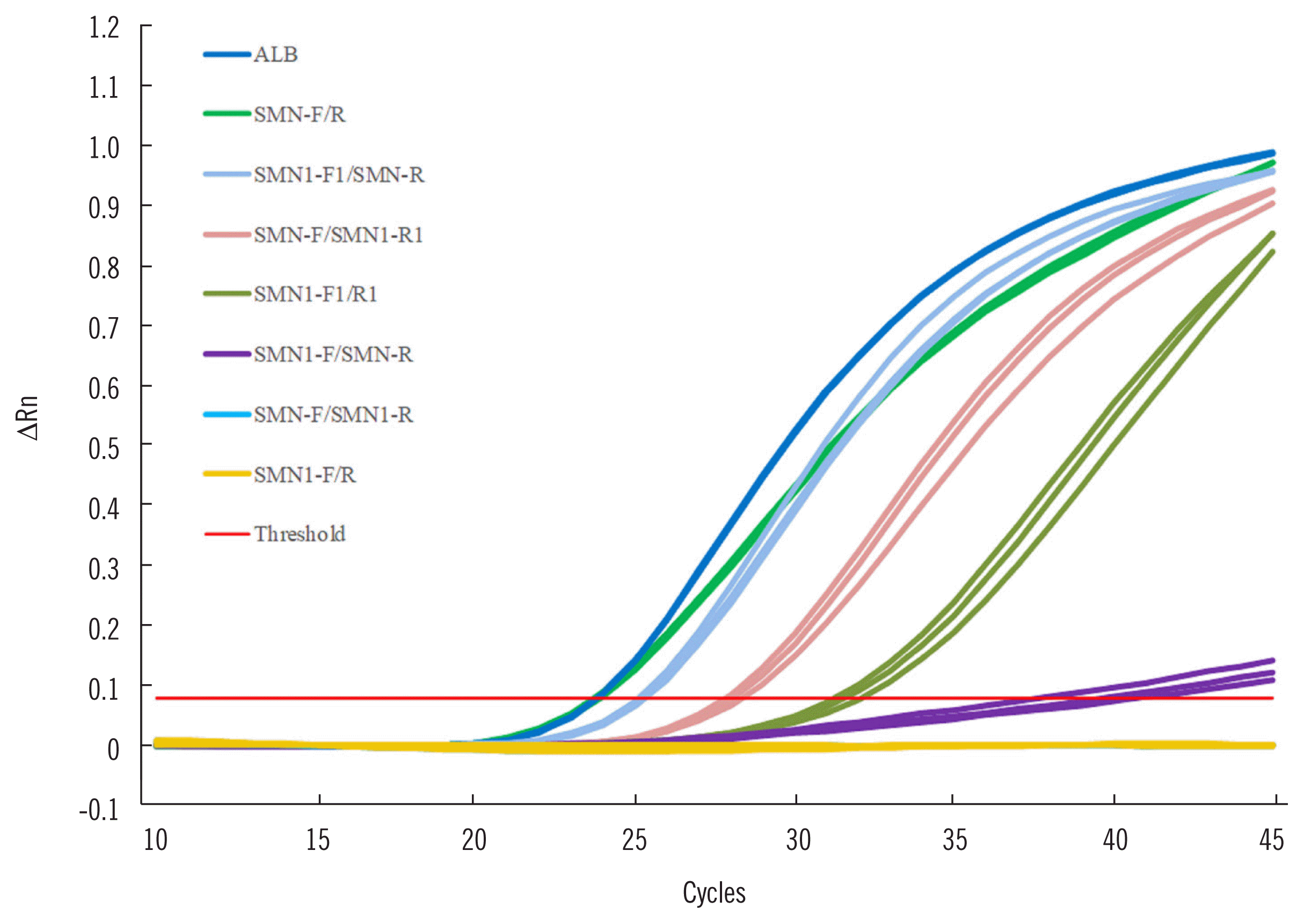
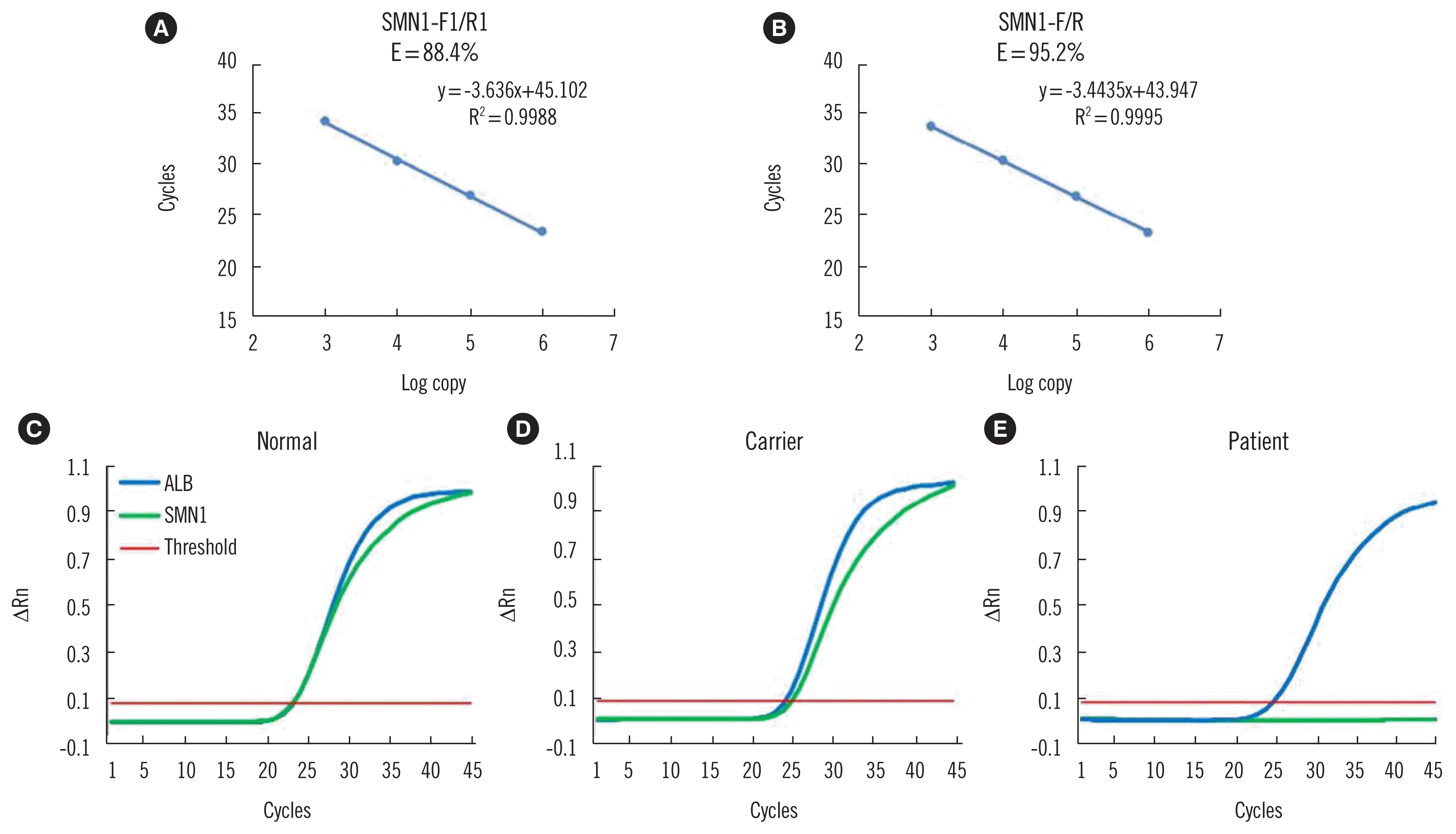




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download