This article has been
cited by other articles in ScienceCentral.
Dear Editor,
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as a novel virus causing pneumonia in December 2019 in Wuhan city, Hubei province, China, which led to a huge outbreak of the disease, now widely known as COVID-19, spreading to 46 countries by 26th February 2020 [
1]. The WHO recommends nucleic acid amplification tests for laboratory confirmation of COVID-19 [
2]. Over nine days, from 18th–26th February 2020, approximately 50,000 tests for suspected cases of COVID-19 were performed in Korea, resulting in a diagnosis of 1,742 new cases in this period [
3,
4]. On 6th February 2020, Emergency Use Authorization (EUA) for COVID-19 testing was implemented in Korea, permitting rapid expansion of capacity in clinical and public health laboratories [
5]. Such a surge in demand for laboratory diagnostic tests inevitably results in insufficient validation of new tests accompanied by lack of resources.
On 17th February 2020, real-time reverse transcription (rRT)-PCR was set up for SARS-CoV-2 detection by Allplex 2019-nCoV assay (Seegene, Seoul, Korea) using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) in the clinical laboratory in Asan Medical Center, Seoul, Korea. However, we experienced cycle threshold (Ct) prolongation of the internal control (IC) and therefore investigated the cause. The Institutional Review Board of Asan Medical Center (IRB No. 2020-0370) exempted the study from further review and the need for informed consent.
Sputum samples collected without preservation and nasopharyngeal (NP) samples collected using flocked swabs were transported in a UTM Viral Transport system (Copan, Brescia, Italy). Each sample was first spiked with the IC in Allplex kit, and then RNA was extracted with the automated silica-coated magnetic bead system of Real-Prep (BioSewoom Inc., Seoul, Korea) using Real-Prep Viral DNA/RNA kit (BioSewoom). This system was first introduced in the laboratory on 14th February 2020 for SARS-CoV-2 rRT-PCR. For rRT-PCR, 8 μL of the RNA extract was manually added to PCR mixtures in Allplex kit, and Ct values of IC were monitored for all samples. Ninety-four runs comprising 3,345 tests were performed over nine days from 18th-26th February 2020. There were no positive clinical samples. The average Ct value of IC was 22.60±0.99 at the first run, which increased to a maximum of 34.97±2.99 among the runs performed on 26th February 2020 for NP samples; a similar increase from 23.94±1.48 to 38.67 was detected for sputum samples (
Fig. 1). There was an increased failure in amplification of IC during the same period, with a maximum failure rate of 41.2% for NP and 88.9% for sputum samples on 26th February 2020 (
Table 1).
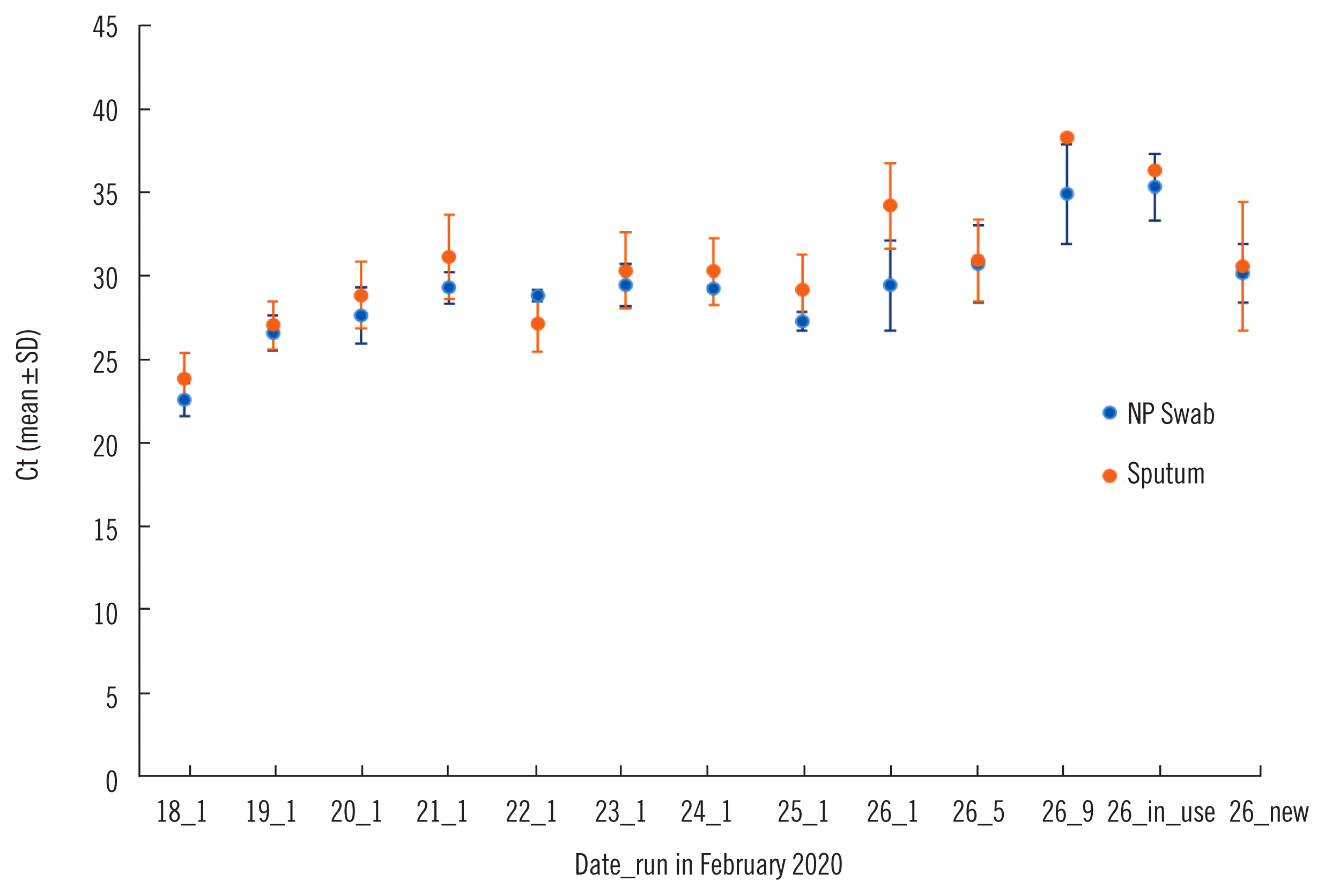 | Fig. 1
Trends of Ct values of the IC in rRT-PCR for SARS-CoV-2 detection. At the end of 26th February 2020, in 31 samples, RNA was extracted using the Real-Prep (BioSewoom, Seoul, Korea) system in use, 2–26 (in use), and in 35 samples, RNA was extracted using the new Real-Prep system, 2–26 (new); all of these were submitted to a single run of PCR for comparison.
Abbreviations: Ct, cycle threshold; IC, internal control; rRT-PCR, real-time reverse transcription PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NP, nasopharyngeal.

|
Table 1
Failure rates of IC for SARS-CoV-2 rRT-PCR for nine days in February 2020
|
Date_run*
|
N failures/N tests (%) |
|
NP swab |
Sputum |
Total |
|
18_1 |
0/13 (0) |
0/11 (0) |
0/24 (0) |
|
19_1 |
0/22 (0) |
0/21 (0) |
0/43 (0) |
|
20_1 |
0/31 (0) |
1/34 (2.9) |
1/65 (1.5) |
|
21_1 |
0/15 (0) |
0/16 (0) |
0/31 (0) |
|
22_1 |
0/24 (0) |
0/5 (0) |
0/29 (0) |
|
23_1 |
0/15 (0) |
1/10 (10) |
1/25 (4) |
|
24_1 |
1/19 (5.3) |
0/13 (0) |
1/32 (3.1) |
|
25_1 |
0/21 (0) |
0/11 (0) |
0/32 (0) |
|
26_1 |
0/20 (0) |
0/17 (0) |
0/37 (0) |
|
26_5 |
19/47 (40.4) |
11/17 (64.7) |
30/64 (46.9) |
|
26_9 |
7/17 (41.2) |
8/9 (88.9) |
15/26 (57.7) |
|
26_in_use*
|
7/21 (33.3) |
9/10 (90) |
16/31 (51.6) |
|
26_new†
|
0/28 (0) |
0/7 (0) |
0/35 (0) |

To evaluate the performance of the RNA extraction step, 31 and 35 samples were prepared using the Real-Prep-in-use and the new Real-Prep system, respectively, and all 66 eluates were submitted to the same run of rRT-PCR. The IC failure rate of NP samples was 51.6% with an average Ct value of 35.4±2.00 using Real-Prep-in-use, but it was 0% with a Ct value of 30.24±1.77 using the new Real-Prep system (
Fig. 1). Examination of the Real-Prep-in-use by the manufacturer revealed that all magnetic rods were coated with a substantial number of magnetic beads, which would hamper the ability to hold magnetic beads. Therefore, Ct prolongation and failure of IC were attributed to the poor quality of the RNA extraction step.
The purpose of IC is to validate nucleic acid extraction and the efficiency of PCR [
6]. For this purpose, housekeeping genes intrinsic to samples are used as IC, or artificial IC can be spiked in the samples before preparation. Therefore, a well-designed IC is an essential component of PCR kits to detect viral target genes [
7]. This is particularly relevant for EUA-approved kits, which are exempt from clinical validation requirements. Although five SARS-CoV-2 rRT-PCR have been approved by the EUA as of 23rd March 2020 in Korea [
8], RNA bacteriophage as an IC is available with the Allplex kit only, monitoring the entire process from RNA extraction to PCR (
http://www.seegene.com/assays/allplex_2019_ncov_assay). The analytical performance of PCR is largely dependent on the quality of the sample and nucleic acid extraction efficiency [
9]. We found more prominent deterioration in PCR efficiency of IC with sputum samples than with NP samples. Both sputum and NP samples were recommended with same cut-off of Ct value <40 by the manufacturer, but sputum samples require homogenization, which critically influences the efficiency to extract nucleic acids and eliminate interferents that can inhibit PCR [
7,
9]. Contamination of magnetic rods is an additional concern that could lead to false positivity, although no positive samples were detected during this investigation.
As the experience of the Middle East Respiratory Syndrome outbreak of 2015 in Korea, an epidemic of emerging infectious diseases necessitates clinical laboratories to conduct new tests at a large scale in a short period of time with kits and equipment that have not been thoroughly validated [
10]. This situation poses a risk affecting the quality of the diagnostic tests approved by the EUA. Accordingly, EUA-approved kits should include adequate ICs to allow for monitoring of the overall analytical performance. Clinical microbiologists should closely monitor the ICs for each run with products under an EUA condition.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download