This article has been corrected. See "Erratum: COVID-19 Molecular Testing in Korea: Practical Essentials and Answers From Experts Based on Experiences of Emergency Use Authorization Assays" in Volume 41 on page 126.
Abstract
Coronavirus disease 2019 (COVID-19) is a respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Early detection of COVID-19 and immediate isolation of infected patients from the naive population are important to prevent further pandemic spread of the infection. Real-time reverse transcription (RT)-PCR to detect SARS-CoV-2 RNA is currently the most reliable diagnostic method for confirming COVID-19 worldwide. Guidelines for clinical laboratories on the COVID-19 diagnosis have been recently published by Korean Society for Laboratory Medicine and the Korea Centers for Disease Control and Prevention. However, these formal guidelines do not address common practical laboratory issues related to COVID-19 real-time RT-PCR testing and their solutions. Therefore, this guideline is intended as a practical and technical supplement to the “Guidelines for Laboratory Diagnosis of COVID-19 in Korea”.
Coronavirus disease 2019 (COVID-19) is a respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], which was first identified in Wuhan, China, in December 2019, and has rapidly spread globally, resulting in the ongoing pandemic [2]. Recently, Korean Society for Laboratory Medicine (KSLM) and the Korea Centers for Disease Control and Prevention (KCDC) published guidelines for diagnosing COVID-19 in clinical laboratories in Korea [3]. These guidelines provide appropriate solutions to COVID-19 diagnosis. However, many practical and technical issues frequently arise in clinical laboratories conducting COVID-19 testing, including those involving nucleic acid extraction, nucleic acid amplification reagents, and interpretation of test results. The COVID-19 Diagnosis Test Management Committee was established by the KCDC on Feb 28, 2020, to provide appropriate solutions to such matters in the field through careful review and counseling by specialized experts in the public and private sectors. On behalf of the COVID-19 Diagnosis Test Management Committee, we present supplementary information to the “Guidelines for Laboratory Diagnosis of COVID-19 in Korea” to provide solutions for practical issues faced when conducting COVID-19 diagnostic testing using real-time reverse transcription (RT)-PCR [3]. These practical guidelines are mostly based on expert opinions regarding samples, nucleic acid extraction, nucleic acid amplification reagents, and interpretation of test results and have been formed with the experience from more than 700,000 COVID-19 tests using five emergency use authorization (EUA) assays in Korea.
This guideline was exempted from review by the Asan Medical Center’s Institutional Review Board, Seoul, Korea, owing to its minimal risk and exemption category fulfillment, including analysis of previously collected information [cycle threshold (Ct) values of samples], without requiring clinical data from other patients [AMC IRB 2020-0548].
In principle, we recommend simultaneous testing of upper and lower respiratory tract samples [3, 4]; however, testing of an upper respiratory tract sample alone may be unavoidable when sputum cannot be obtained [3]. In case of upper respiratory tract samples, several guidelines recommend using a universal transport medium (UTM) for nasopharyngeal (NP) and oropharyngeal (OP) swabs to increase test sensitivity [3–5]. Since several flocked swab products have one swab and one UTM tube, two sets of swabs should be used to place both NP swab and OP swab in the same UTM. Considering the current short supply of flocked swabs, using only the NP swab can be considered. NP swabs resulted in a higher positive rate than OP swabs [6]. Further, our data showed lower Ct values in NP swabs than in OP swabs (Table 1).
FLOQSwab (Copán, Brescia, Italy) in eNAT (Copán) and UTM (Copán) is a flocked swab available in Korea that is easy to use because it has a red line at the molded break point (Fig. 1A). Since eNAT contains guanidine thiocyanate (a type of chaotropic salt) that prevents the degradation of nucleic acids in the medium, it is a good option when testing microbial nucleic acids only; the same applies to the COVID-19 real-time RT-PCR test. HydraFlocked (Diagnostic Hybrids, Athens, OH, USA) and NFS-Swab Applicator (Noble Bio, Hwaseong, Korea) do not include a red line at the molded break point (Fig. 1A). In addition, it is difficult to break the shaft of an NFS-Swab Applicator. The product manual recommends inserting the shaft groove between the lid and the rim of the tube to facilitate breaking (Fig. 1B). When using REST UTM (Noble Bio), it is important to vortex the sample thoroughly to remove cells from the swab, since the swab does not contain any beads.
The best-known method for sputum homogenization for Middle East respiratory syndrome virus tests is proteinase K and DNase I treatment [7]. The optimal temperature for proteinase K activity ranges from 50°C to 65°C. However, heat inactivation of samples through treatment at 56°C for 30 minutes was shown to adversely affect the efficiency of RT-PCR for SARS-CoV-2 detection [8]; therefore, proteinase K treatment at 50–65°C is not recommended for COVID-19 real-time RT-PCR. Use of N-acetyl-L-cysteine dissolved in a sodium citrate solution is also a feasible option for sputum homogenization although the prepared solution is valid for only 24 hours. Caution is required to avoid excessive dilution of the sample due to the addition of a large amount of phosphate-buffered saline (PBS) during homogenization. Good results can be expected by adding 500 μL of sputum to a 2-mL microtube (using a swab), mixing with the same amount of PBS or UTM with glass beads, sufficiently vortexing and centrifuging, and then using only the supernatant for extraction. A sputum sample mixed and homogenized with the same amount of Liquillizer (MetaSystems, Altlussheim, Germany) can also be used for molecular diagnosis and sputum culture. Although access to the Liquillizer reagent is hindered owing to its limited distribution in Korea, it has excellent mucolytic and homogenizing performance.
Before selecting or using a reagent for nucleic acid extraction in a laboratory, it is important to determine its performance in extracting nucleic acids from respiratory tract samples. We recommend comparing the performance of the proposed extraction reagent with that of the reagent from the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Nucleic acids should be extracted from more than 30 respiratory virus samples, including 10 or more respiratory virus-positive samples, using the proposed extraction reagent simultaneously with the QIAamp Viral RNA Mini Kit extraction reagent. Respiratory virus multiplex PCR should then be performed, and the positive rate of extraction and Ct values of the target viral gene and internal control (IC) for the proposed reagent should be compared with those for the QIAamp Viral RNA Mini Kit extraction reagent. Although there are no definitive criteria for assessment, a reagent is generally considered comparable in performance to that of the QIAamp Viral RNA Mini Kit extraction reagent, if the Ct value difference is ≤1.66 of 0.5 log10 copies/mL equivalents [9,10].
Reagent supply stability should be considered when using imported reagents, such as MagNA Pure 96 (Roche Molecular Systems, Pleasanton, CA, USA), NucliSens easyMAG (bioMérieux, Marcy l’Etoile, France), and Qiagen EZ1 Advanced XL (Qiagen).
As of May 18, 2020, six real-time RT-PCR assays have been approved in Korea under EUA (Table 2). The PowerChek 2019-nCoV kit (Kogenebiotech, Seoul, Korea) uses two PCR tubes per sample, while the other assays require only one PCR tube per sample. The product manual of the DiaPlexQ 2019-nCoV kit (SolGent, Daejeon, Korea) states that a sample can indicate COVID-19 infection if the result for one of the two genes is positive. This is in contrast with the KSLM guidelines, which specifies that confirmation of COVID-19 requires positive results for all genes [3]. Since the Real-Q 2019-nCoV kit (BioSewoom, Seoul, Korea) and BioCore 2019-nCoV Real Time PCR kit (BioCore, Seoul, Korea) use the human nuclear RNase P and human β-globin genes as the IC, the Ct value of the IC is increased proportionally to the dilution factor, if the sample or RNA is diluted for retesting. As KSLM and the KCDC have only evaluated the CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, USA) and the Applied Biosystems 7500 FAST and 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) for six EUA assays [11], performance of the device intended for use should be compared.
To change or add EUA assays, parallel tests using at least 10 positive and 10 negative samples should be performed, and the results should be reviewed by the person in charge of the laboratory. The fourth external quality assessment by the Korean Association of External Quality Assessment Service used an inactivated culture of SARS-CoV-2 [12]. This sample can be used for parallel tests.
Both Allplex 2019-nCoV (Seegene, Seoul, Korea) and Standard M nCoV Real-Time Detection kits (SD Biosensors, Suwon, Korea) have the advantage of facilitating the observation of the entire process of COVID-19 molecular testing to check nucleic acid extraction, the impact of the PCR inhibitor, and nucleic acid amplification since the IC is directly added to the sample before nucleic acid extraction. It is not possible to check the nucleic acid extraction with other assays in that the IC is added directly into the PCR mixture (Table 2).
When using a nucleic acid extraction reagent that requires the sample to be suspended in lysis buffer, the sample should be added to the lysis buffer prior to the IC. Although sodium dodecyl sulfate, a detergent in the lysis buffer, has almost no impact on nucleic acids (DNA and RNA), since it has a negative charge, we recommend following the above procedure when adding a small amount of the IC in a large volume of lysis buffer to prevent possible damage of the bacteriophage or lentivirus RNA. Further proper addition of the IC to the sample should also be confirmed, since 5–10 μL is a minor amount. If the IC is placed at the top (rim) of a reaction tube (or reaction well), amplification failure may occur due to inadequate mixing with the sample. The IC of the “Positive Control” of Allplex 2019-nCoV kit is plasmid DNA. The deviation of the Ct value from the IC of the “Positive Control” can be large when using immediately thawed reagent. Therefore, it is necessary to extract and test the sample again if there is no amplification of the IC. If there is still no amplification of the IC after re-extraction and retesting, we recommend performing a retest using a separate sample. When obtaining another sample is not feasible, the possibility of the presence of PCR inhibitors in the extracted nucleic acid solution should be ruled out using the current testing material. We recommend diluting the extracted nucleic acids by a factor of 10 in a retest in this case.
It is possible to improve the test sensitivity by increasing the amount of the “input” RNA using the extracted RNA instead of “DNase-, RNase-free distilled water” to reach the final PCR volume required. However, we do not recommend this option since this also typically results in the addition of more PCR inhibitors. As mentioned above, a 10-fold dilution of the RNA can be considered in a retest, if the IC is not amplified.
In the case of different results between upper and lower respiratory tract samples, the first step would be to check for any clerical error (e.g., misidentification of patient name or hospital identification number, labeling error, or switched samples). In addition, the adequacy of the two samples from the patient should be considered; it is reasonable to presume that the sputum may not be “appropriate.” Moreover, the level of pre-treatment (mucolytic and homogenization) of the sputum should be considered. Infection can be confirmed with a positive result from either sample when the criteria described below are met.
The two SARS-like beta coronaviruses (sarbecoviruses) that infect humans are the SARS-CoV-2 strain that is currently active and SARS-CoV that emerged in 2003 [13]. There is no evidence that SARS-CoV has reappeared. The E gene is not amplified from beta coronaviruses (e.g., OC43 and HKU1) other than sarbecoviruses [13]. Therefore, if the test shows an E-positive and RdRp-negative result, the position of the E gene reaction well (PCR tube) should be checked for its proximity to the positive control well, which could suggest the possibility of cross-contamination; in this case, extraction and retesting of the sample is necessary. This situation is applicable only to newly diagnosed patients (first testing of the first sample). Only one of the target genes (E gene, RdRp gene, and N gene) may show positivity during the follow-up test of a confirmed case.
When using the Allplex 2019-nCoV kit, if the Seegene Viewer shows positive Ct values in the 20s for the E gene only, the amplification curve should be evaluated, separately selecting the 6-carboxyfluorescein (6-FAM) channel. A nonspecific reaction curve will appear, similar to the graph shown in Fig. 2.
One of the possible causes for negative control amplification is template contamination from the environment or amplicon [14]. To check the source, a wipe test of the biosafety cabinet (BSC), micropipette, extraction device, PCR workstation, and PCR device that handles the samples should be performed [15]. Or alternatively, a flocked swab for NP swab collection could be used for the surface of the device equivalent to a sheet of A4 paper. The surface should be scraped to collect the sample and then be tested in the same manner as that used for testing a patient’s sample. Another possible cause for negative control amplification is random nonspecific amplification or probe instability. In that case, a similar nonspecific amplification would occur in the patient’s sample reaction well, and retesting would often show a clean negative result.
The currently available assays have been approved for emergency use; therefore, the results obtained using them require careful interpretation. Multiple genes from the same sample should be detected (i.e., simultaneous detection of the E gene and RdRp gene) and analyzed with another sample collected from the same patient (i.e., the sputum and NP swab together). Consider re-extraction and retesting, particularly using another sample for hospitalized patients.
With the Allplex 2019-nCoV kit, the Ct value of the E gene in a positive sample is 1.2–1.5 cycles lower on average than that of the RdRp gene, and the Ct value of the RdRp gene is approximately 1.5 cycles lower on average than that of the N gene. For samples with a high viral load, it is helpful to confirm, if the Ct values of the individual genes are observed in the expected order. With the PowerChek 2019-nCoV kit, the E gene Ct value of the positive sample is lower than that of the RdRp gene, and the difference between the Ct values of these genes is approximately 2.5. However, with the Standard M nCoV Real-Time detection kit, the RdRp gene Ct value is lower by approximately 1.5 cycles than that of the E gene Ct value (data not shown). SARS-CoV-2 infection can be confirmed without retesting when the Ct values of the E and RdRp genes are 33.5 cycles or lower. However, checking the position of the reaction well (PCR tube) near the positive control is also necessary to eliminate the possibility of cross-contamination and verify that the amplification sequence of the E, RdRp, and N genes follows that order (in the case of the Allplex 2019-nCoV kit).
The theoretical Ct value to amplify one copy of a target gene using a 20-μL PCR is 37 [16]. However, the Ct value corresponding to one-copy amplification may differ in actual samples due to various factors, such as the presence of PCR inhibitors in the sample, activity of the one-step RT-PCR enzyme, and the fluorescence sensitivity of the device. In the case of the PowerChek 2019-nCoV kit, the RdRp gene may show an amplification curve at the end of the amplification stage (upward drift). Such an upward drift of the RdRp gene without E gene amplification indicates a likely negative result. In that case, re-extraction and retesting should be considered. If obtaining another sample is not possible, a common situation for many referral laboratories, we recommend a retest when the Ct value of either the E or the RdRp gene is higher than 33.5 and below 40 (the gray zone suggested by the Committee).
A negative result can be confirmed if the retest result is negative, and a positive result can be confirmed if the retest result shows that the Ct values of both the E and RdRp genes are 33.5 or lower. A result of “indeterminate” should be reported followed by a retest with another sample, if the retest result of the sample is “indeterminate.”
Table 3 shows the daily test results of a patient who was confirmed to be infected with SARS-CoV-2 using the Allplex 2019-nCoV kit, as an example for follow-up to make a decision for release from quarantine (note that this does not represent a typical case).
When retesting a follow-up sample, some genes are often detected and some are not, and the types of detected genes often change (Table 3). For example, a retest of a sample that was positive for both the E and N genes in the first test may show that the sample is positive for the RdRp and N genes in the follow-up. Based on the experience from hospitals with many confirmed patients, the N gene appears to remain detectable longer than other genes in many cases. Therefore, it is important to interpret the results carefully when a sample from a confirmed patient is being tested for deciding on quarantine release. Avoiding simple application of the manufacturers’ suggested interpretation criteria is strongly advised. We recommend reporting the result as “positive” regardless of the Ct value if all target genes of the kit are amplified in the follow-up sample of a confirmed patient. An “Indeterminate (inconclusive)” result should be reported, if one or more, but not all genes included in the kit show an amplification curve after the cut-off when using a follow-up sample. If the amplification curve shows an exponential function, an “indeterminate” result should be reported, along with an explanation to clinicians that this does not mean “negative.” This should be explicitly stated in the report in the case of a confirmed patient’s sample being tested at a referral laboratory.
The KCDC’s criteria for the quarantine release of patients with COVID-19 include clinical improvement with negative results from at least two consecutive respiratory samples collected ≥24 hours apart [17]. As for the United States Centers for Disease Control and Prevention, the test-based criteria for discontinuation of transmission-based precautions is the resolution of fever without the use of antipyretic drugs, improvement of respiratory symptoms, and negative results for SARS-CoV-2 RNA from at least two consecutive respiratory samples collected ≥24 hours apart [18]. Thus, two consecutive negative results from respiratory samples and clinical judgement are the minimum requirements for quarantine release.
Several EUA real-time RT-PCR assays for SARS-CoV-2 have enabled rapid diagnosis of COVID-19 with subsequent improvement in infection control. However, proper testing and interpretation of results require understanding the strengths and limitations of EUA assays. Results of molecular diagnostic testing for COVID-19 should be interpreted in the context of the individual case presentation, clinical illness, and epidemiological correlations.
ACKNOWLEDGEMENTS
The authors extend their gratitude to Dr. John Jeongseok Yang for English proofreading and feedback and to Dr. Chang Kyu Lee for sharing valuable results regarding sputum homogenization.
Notes
REFERENCES
1. World Health Organization. Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (Updated on February 11, 2020).
2. World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (Updated on March 11, 2020).
3. Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020; 40:351–60.

4. Centers for Disease Control and Prevention. Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19). https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html (Updated on April 14, 2020).
5. World Health Organization. Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance (Updated on March 2, 2020).
6. Wang X, Tan L, Wang X, Liu W, Lu Y, Cheng L, et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis. 2020; 94:107–9.

7. Sung H, Yong D, Ki CS, Kim JS, Seong MW, Lee H, et al. Comparative evaluation of three homogenization methods for isolating Middle East respiratory syndrome coronavirus nucleic acids from sputum samples for real-time reverse transcription PCR. Ann Lab Med. 2016; 36:457–62.

8. Pan Y, Long L, Zhang D, Yan T, Cui S, Yang P, et al. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. 2020; Apr. 4. pii: hvaa091. DOI: 10.1093/clinchem/hvaa091.

9. Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, et al. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996; 34:2695–701.

10. Kim S, Park SJ, Namgoong S, Sung H, Kim MN. Comparative evaluation of two automated systems for nucleic acid extraction of BK virus: NucliSens easyMAG versus BioRobot MDx. J Virol Methods. 2009; 162:208–12.

11. COVID-19 Diagnosis Test Management Committee. Q&A for COVID-19 testings. http://www.kslm.org/ (Updated on March 24, 2020).
12. Sung H, Yoo CK, Han MG, Lee SW, Lee H, Chun S, et al. Preparedness and rapid implementation of external quality assessment helped quickly increase COVID-19 testing capacity in the Republic of Korea. Clin Chem. 2020; Apr. 22. pii: hvaa097. DOI: 10.1093/clinchem/hvaa097.

13. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020; 25:2000045.

14. CLSI. Establishing molecular testing in clinical laboratory environments; approved guideline. CLSI MM19-A. Wayne, PA: Clinical and Laboratory Standards Institute;2011.
15. Park GW, Chhabra P, Vinjé J. Swab sampling method for the detection of human norovirus on surface. J Vis Exp. 2017; 120:55205.
16. Thermo Fisher Scientific. Distinguishing-real-signal-from-background-noise-ask-taqman-41. https://www.thermofisher.com/blog/behindthebench/ (Updated on December 23, 2016).
17. Korea Centers for Disease Control and Prevention. Management guidelines for coronavirus disease-19. Version 8. http://ncov.mohw.go.kr/duBoardList.do?brdId=2&brdGubun=28 (Updated on May 11, 2020).
18. Centers for Disease Control and Prevention. Criteria for return to work for healthcare personnel with suspected or confirmed COVID-19 (Interim Guidance). https://www.cdc.gov/coronavirus/2019-ncov/hcp/return-to-work.html#f1 (Updated on April 30, 2020).
Fig. 1
Swab products available in Korea and how to use them. (A) Commercial flocked swabs and universal transport media (UTM). eNAT and FLOQSwab (Copán, Brescia, Italy), UTM and FLOQSwab (Copán), UTM (Copán) and HydraFlocked (Diagnostic Hybrids, Athens, OH, USA), and REST UTM and NFS-Swab Applicator (Noble Bio, Hwaseong, Korea) (left to right). (B) Breaking swab shaft between lid and upper rim of UTM.
Abbreviation: UTM, universal transport medium.
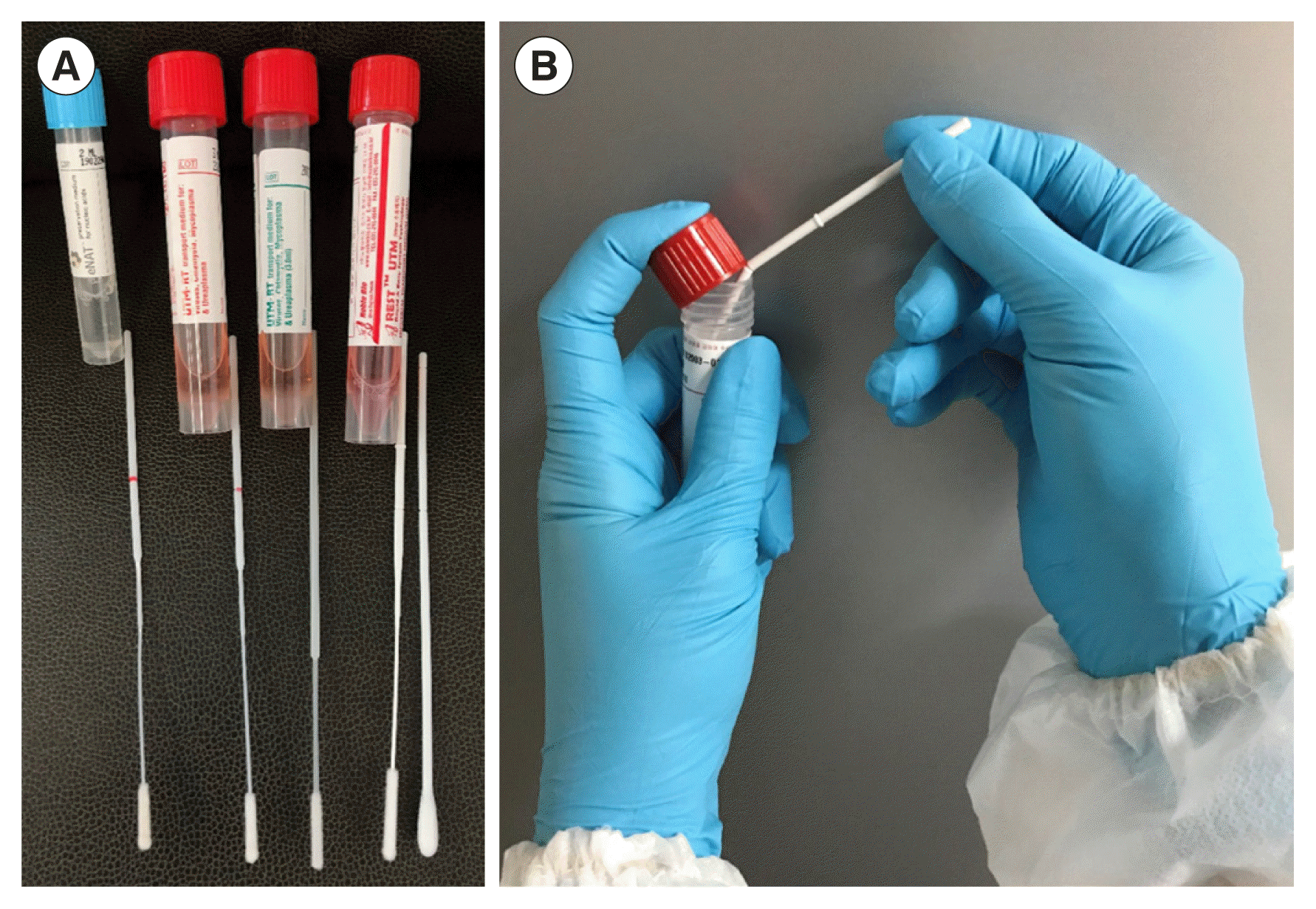
Fig. 2
Nonspecific E gene amplification using Allplex 2019-nCoV assay. (A) Raw data obtained using Seegene Viewer (Seegene, Seoul, Korea). (B) Raw data obtained using CFX96 Real-Time Detection System. Circles show non-exponential noise curves.
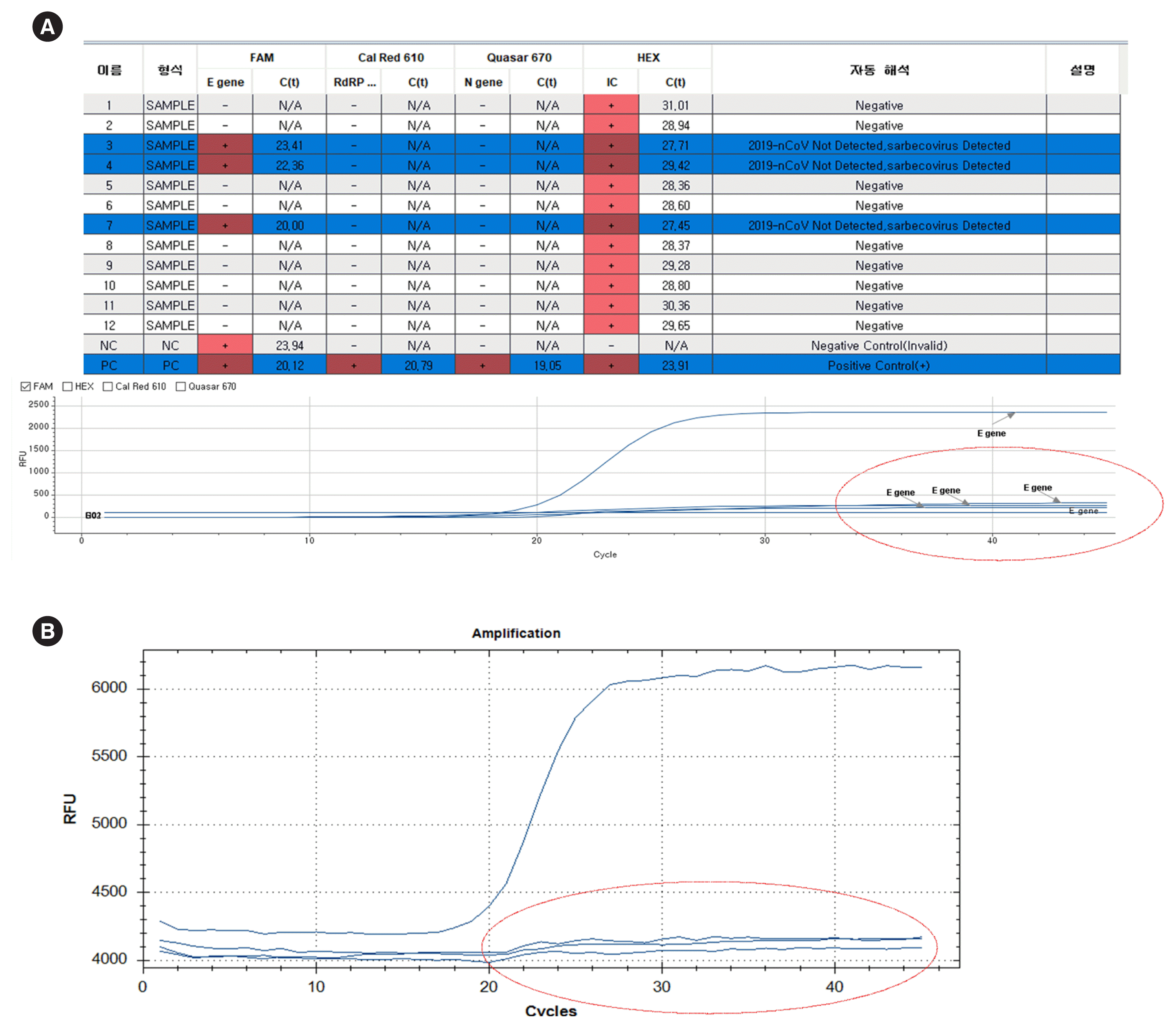
Table 1
Cycle threshold values of nasopharyngeal swabs and oropharyngeal swabs from five COVID-19 patients, tested using the Allplex 2019-nCoV kit (Seegene, Seoul, Korea)
Table 2
The characteristics of six EUA real-time RT-PCR assays for COVID-19 diagnosis
This table was modified from Table 1 by Hong, et al. [3] with permission from Annals of Laboratory Medicine.
Table 3
Representative serial results of cycle threshold values from SARS-CoV-2 real-time RT-PCR assay
| Hospital day | Sputum | Nasopharyngeal swab | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| E gene | RdRp gene | N gene | Interpretation | E gene | RdRp gene | N gene | Interpretation | |
| 9 | 22.61 | 24.68 | 26.24 | Positive | 32.78 | 34.50 | 34.06 | Positive |
|
|
||||||||
| 10 | ND | 38.70 | 37.35 | Indeterminate | 34.09 | 36.79 | 35.81 | Positive |
|
|
||||||||
| 11 | 35.42 | ND | 37.13 | Indeterminate | ND | ND | ND | Negative |
|
|
||||||||
| 12 | ND | ND | ND | Negative | ND | ND | ND | Negative |
|
|
||||||||
| 13 | ND | 39.14 | ND | Indeterminate | ND | ND | ND | Negative |
|
|
||||||||
| 14 | ND | ND | ND | Negative | ND | ND | ND | Negative |
|
|
||||||||
| 15 | ND | ND | 38.42 | Indeterminate | ND | ND | 38.56 | Indeterminate |
|
|
||||||||
| 16 | 33.60 | 37.34 | 38.94 | Positive | ND | 37.64 | 36.51 | Indeterminate |
|
|
||||||||
| 17 | ND | 35.65 | ND | Indeterminate | ND | ND | 38.56 | Indeterminate |
|
|
||||||||
| 18 | ND | ND | ND | Negative | 34.63 | 32.71 | 35.39 | Positive |
|
|
||||||||
| 19 | ND | ND | ND | Negative | ND | ND | 39.16 | Indeterminate |
|
|
||||||||
| 20 | 32.48 | ND | 34.11 | Indeterminate | ND | ND | ND | Negative |
|
|
||||||||
| 21 | ND | ND | ND | Negative | ND | ND | ND | Negative |
|
|
||||||||
| 22 | ND | ND | ND | Negative | ND | 37.60 | ND | Indeterminate |
|
|
||||||||
| 23 | NT | NT | NT | - | ND | ND | ND | Negative |
|
|
||||||||
| 24* | NT | NT | NT | - | ND | ND | ND | Negative |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download