Dear Editor,
Coprobacillus catenaformis is a gram-positive, obligatorily anaerobic, non-spore-forming rod bacteria. In 2000, Coprobacillus gen. nov. (Copro, feces; bacillus, a small rod) catenaformis spp. nov. (catena, chain; formis, shaped) was proposed as a member of the Clostridium subphylum of gram-positive bacteria that is phylogenetically closely associated with Clostridium ramosum and C. spiroforme [1]. C. catenaformis, the only known species in the genus Coprobacillus, is a member of the intestinal microbiota of humans and mice, as shown by metagenomics studies [2, 3]. To the best of our knowledge, this species has not been isolated from clinical specimens, including blood cultures. We report the first case of bacteremia caused by C. catenaformis. The Institutional Review Board of Samsung Medical Center, Seoul, Korea, approved this study (approval number: 2020-03-085) and waived the need for informed consent.
A patient in her 60s, who received several cycles of chemotherapy and radiotherapy for non-small cell lung cancer with pleural and bone metastases since August 2014, was admitted to Samsung Medical Center in August 2019 for a decompressive laminectomy with pedicle screw fixation and tumor removal. On post-operative day 8, a bisacodyl enema was administered for >10 days without defecation. A few hours after the enema, the patient complained of chilling sensation and had a body temperature of 38.6°C. Her blood pressure was 108/55 mm Hg, pulse rate was 142 bpm, and respiratory rate was 18 breaths per minute. Her C-reactive protein was 56.3 (reference interval, 0–5.0) mg/L and leukocyte count was 10.44×109/L (neutrophils, 98.0%). After preparing two sets of aerobic and anaerobic blood culture bottles (bioMérieux, Marcy l’Etoile, France), empirical antibiotic therapy with intravenous piperacillin/tazobactam 4.5 g at eight hours interval was administered. On day 2 of incubation in a BacT/ALERT 3D blood culture instrument (bioMérieux), growth was observed in both anaerobic bottles of the two sets of blood cultures. Signal-positive bottles were subcultured and the following bacteria were isolated: Eubacterium callanderi and unidentified gram-positive bacilli from both sets of blood culture bottles and Granulicatella elegans from the blood culture bottle of the first set.
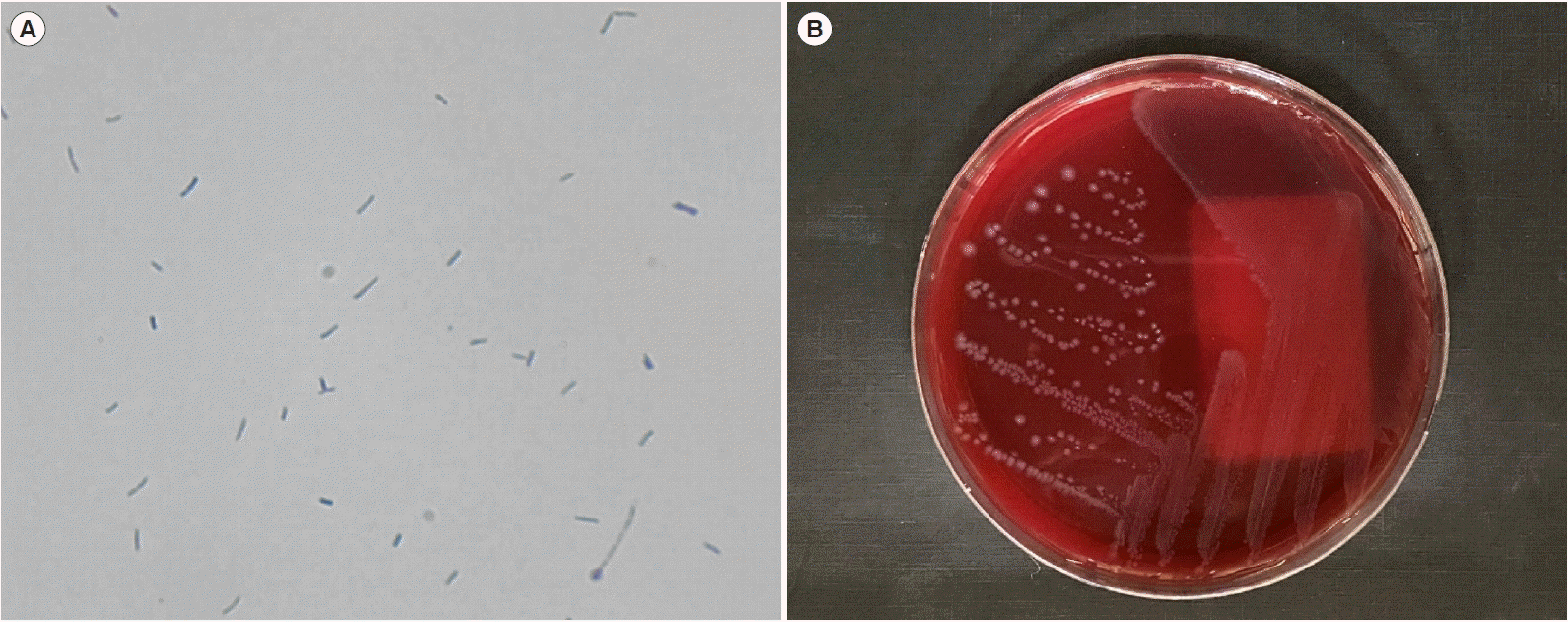
Microscopic examination revealed that the unidentified species was a gram-positive, non-spore-forming bacillus, which grew as small, grayish colonies on a Brucella agar plate (Fig. 1). The microorganism could not be identified by matrix assisted laser desorption ionization-time of flights mass spectrometry using the VITEK-MS (bioMérieux) or the GP card of the VITEK 2 System (bioMérieux). To accurately identify the isolate, we performed 16S rRNA target sequencing using the universal primers described in the Clinical and Laboratory Standards Institute guidelines [4]. The amplified sequences were compared with sequences in the GenBank database using the basic local alignment search tool algorithm. The 16S rRNA sequence of the isolate exhibited 99.17% (715/721 bp) similarity with C. catenaformis (GenBank accession number AB030219.1). The second-best match was Longibaculum muris, with 92.18% (672/729 bp) similarity. When the sequence was submitted to the Ez-Taxon database (http://www.ezbiocloud.net), the best-matched strain was C. catenaformis (99.86%), which was accepted as the final confirmation of identity.
The patient was successfully treated with a 14-day course of intravenous piperacillin/tazobactam. Follow-up blood cultures from the peripheral veins on days 2, 5, and 14 were negative; the patient’s condition improved and he/she was transferred to a long-term care facility.
To date, several studies have elucidated the roles of Coprobacillus species in the intestinal microbiota. In the human intestinal microbiota, the abundance of Coprobacillus increases with biological age [3] and is diminished in patients with multiple sclerosis [5]. In a mouse experiment, Coprobacillus played a role in inhibiting other microorganisms in the intestinal microbiota, including Clostridioides difficile [2]. To the best of our knowledge, this is the first case demonstrating the possibility of opportunistic Coprobacillus infections. Our patient had a polymicrobial bloodstream infection caused by C. catenaformis, E. callanderi, and G. elegans.
Eubacterium and Granulicatella species are present in the human mouth and intestinal tract and have been implicated as the cause of bacteremia [6–8]. The most common source of polymicrobial bloodstream infections is intra-abdominal infection [9]. The loss of intestinal barrier function contributes to systemic infection, and increased gut permeability is associated with the development of multi-organ dysfunction syndrome [10].
In conclusion, we report the first case of bacteremia caused by Coprobacillus. Although the information regarding human infections of Coprobacillus is limited, Coprobacillus should be considered a possible pathogen, especially in patients at high risk of gastrointestinal infections such as mechanical intestinal injury. Our report emphasizes the importance of Coprobacillus as a potential human pathogen.
Notes
AUTHOR CONTRIBUTIONS
All authors have accepted their responsibility for the entire content of this manuscript and approved submission.
Go to : 
REFERENCES
1. Kageyama A, Benno Y. Coprobacillus catenaformis gen. nov., sp. nov., a new genus and species isolated from human feces. Microbiol Immunol. 2000; 44:23–8.
2. Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, et al. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013; 9:e1003388.

3. Maffei VJ, Kim S, Blanchard Et 4th, Luo M, Jazwinski SM, Taylor CM, et al. Biological aging and the human gut microbiota. J Gerontol A Biol Sci Med Sci. 2017; 72:1474–82.

4. CLSI. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. 2nd ed.CLSI MM18. Wayne, PA: Clinical Laboratory Standards Institute;2018.
5. Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016; 6:28484.

6. Wade WG. The genus Eubacterium and related genera. Dworkin M, Falkow S, editors. The prokaryotes. New York: Springer;2006. p. 823–35.
8. Lee MR, Huang YT, Liao CH, Chuang TY, Wang WJ, Lee SW, et al. Clinical and microbiological characteristics of bacteremia caused by Eggerthella, Paraeggerthella, and Eubacterium species at a university hospital in Taiwan from 2001 to 2010. J Clin Microbiol. 2012; 50:2053–5.
Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download