INTRODUCTION
MATERIALS AND METHODS
Study design
Antimicrobial susceptibility testing (AST)
WGS
AMR genotyping
MLST
CPS genotyping
Virulence gene profiling
Phylogenetic tree analysis
Statistical analysis
RESULTS
Table 1
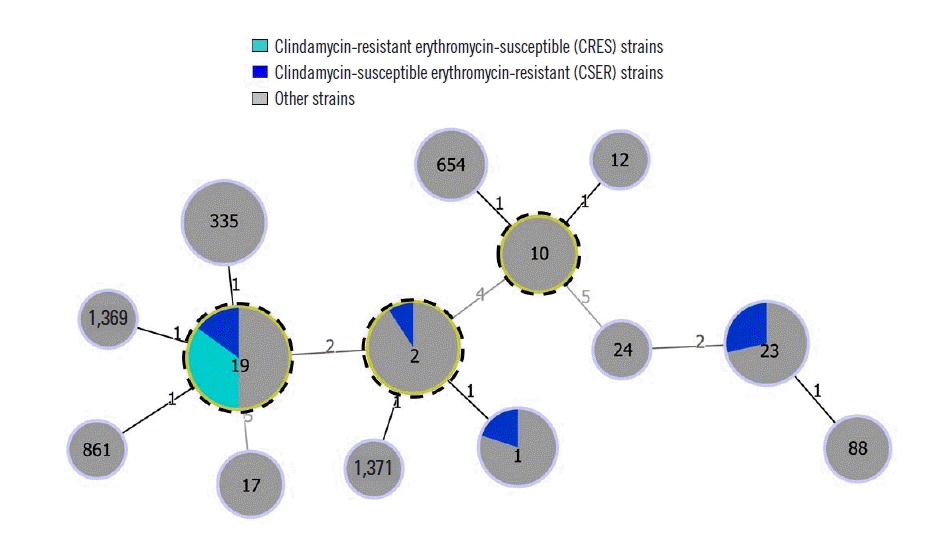 | Fig. 1goeBURST diagram of the relationships among STs of 66 S. agalactiae isolates. The numbers in the circles indicate the STs, and the numbers near the lines indicate the number of differing alleles between the two connected STs. Putative CCs are identified by an outer dotted frame and correspond to the STs with the highest number of single locus variants.
Abbreviations: CC, clonal complex; ST, sequence type.
|
Table 2
| Isolate | Antimicrobial susceptibility pattern | Macrolide/lincosamide resistance gene | Tetracycline resistance gene | Aminoglycoside resistance gene | ST | CPS genotype | Virulence gene profile |
|---|---|---|---|---|---|---|---|
| GCH2 | CRER | erm(B), lnu(B), lsa(E), mre(A) | tet(O) | ant(6)-Ia, ant(6)-Ia, aph(3′)-III | 12 | Ib | bca-bac*-lmb-cylE |
| GCH4 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH5 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH7 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH8 | CSES | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH9 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH10 | CIES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH11 | CSES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH13 | CSES | mre(A) | 1,371 | VIII | rib-lmb-cylE | ||
| GCH14 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH15 | CSES | mre(A) | 2 | VIII | rib-cylE | ||
| GCH16 | CSES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH18 | CSES | lsa(C), mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH19 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH21 | CREI | lsa(C), mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH22 | CSES | mre(A) | 88 | II | lmb-cylE | ||
| GCH25 | CSES | mre(A) | 1 | VI | bca-lmb-cylE | ||
| GCH26 | CSES | mre(A) | tet(O) | 2 | VIII | rib-lmb-cylE | |
| GCH28 | CSES | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH29 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH30 | CRER | erm(B), mre(A) | tet(M) | 1 | V | lmb-cylE | |
| GCH32 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH33 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH34 | CSES | mre(A) | tet(M) | 24 | Ia | bca-lmb-cylE | |
| GCH35 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH36 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH37 | NA | mre(A) | 88 | Ia | lmb-cylE | ||
| GCH38 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 1,369 | III | bca-lmb-cylE |
| GCH39 | CSES | mre(A) | tet(M) | 17 | III | rib-cylE | |
| GCH40 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH41 | CRER | erm(B), mre(A) | tet(O) | ant(6)-Ia, ant(6)-Ia, aph(3′)-III | 17 | III | rib-cylE |
| GCH42 | CSES | mre(A) | 2 | VIII | rib-cylE | ||
| GCH43 | CREI | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH44 | CSEI | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH45 | CSER | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH46 | CSES | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH47 | CRER | erm(A), lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | V | cylE |
| GCH48 | CRER | erm(A), mre(A) | tet(M) | 1 | V | lmb-cylE | |
| GCH49 | CRER | lsa(C), mef(A), mre(A), msr(D) | tet(O) | 861 | III | rib-lmb-cylE | |
| GCH50 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH51 | CRER | erm(B), mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH53 | CSES | mre(A) | tet(M) | 1 | II | lmb-cylE | |
| GCH54 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH55 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH56 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH57 | CRER | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH58 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH59 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH60 | CSES | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH61 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH62 | CRES | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH63 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH64 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH65 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH66 | CRES | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH67 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH68 | CSES | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH70 | CSES | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH71 | CSES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH72 | CSES | erm(B)*, mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH73 | CSER | mre(A) | 1 | VI | bca-lmb-cylE | ||
| GCH74 | CSER | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH75 | CSER | mef(A), mre(A), msr(D) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH76 | CSER | erm(A), mre(A) | tet(M) | 19 | V | lmb-cylE | |
| GCH77 | CSER | mef(A), mre(A), msr(D) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH78 | CSER | erm(B)*, mef(A), mre(A), msr(D) | tet(M) | ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
Abbreviations: AMR, antimicrobial resistance; CPS, capsular polysaccharide; CRER, clindamycin-resistant erythromycin-resistant; CSES, clindamycin-susceptible erythromycin-susceptible; CIES, clindamycin-intermediate erythromycin-susceptible; CREI, clindamycin-resistant erythromycin-intermediate; CSEI, clindamycin-susceptible erythromycin-intermediate; CSER, clindamycin-susceptible erythromycin-resistant; CRES, clindamycin-resistant erythromycin-susceptible; ST, sequence type.
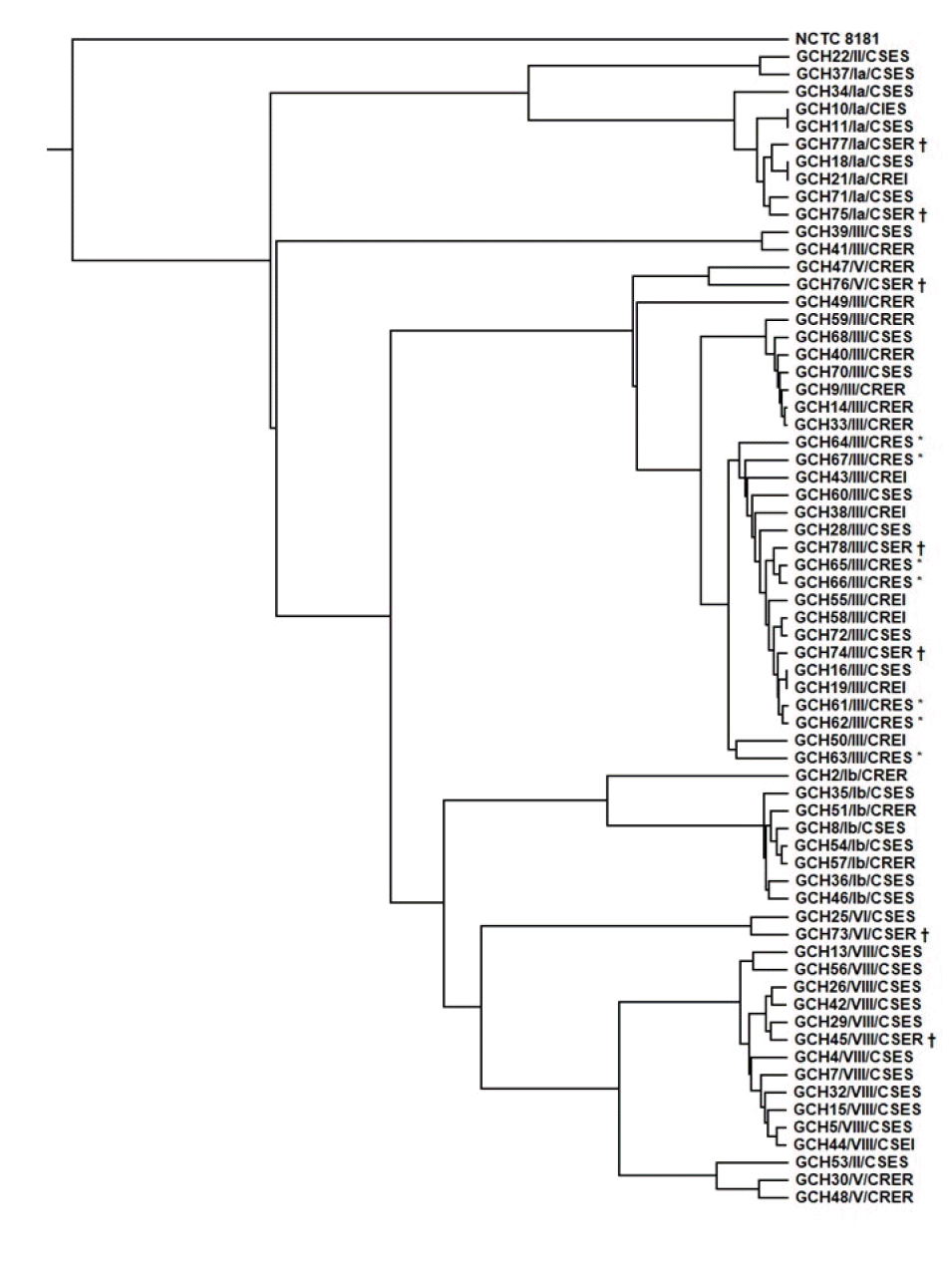 | Fig. 2Phylogenetic tree of 66 S. agalactiae isolates. The phylogenetic tree was constructed using OAT, based on the OrthoANI algorithm, with S. agalactiae strain NCTC8181 (accession number UAVB00000000) as a reference. Asterisks (*) indicate CRES isolates, daggers (†) indicate CSER isolates. There was a concordance of the group distribution on the tree with the CPS genotype distribution (Ia, Ib, III, V, and VIII).
Abbreviations: CPS, capsular polysaccharide; CRES, clindamycin-resistant erythromycin-susceptible; CSER, clindamycin-susceptible erythromycin-resistant; CRER, clindamycin-resistant erythromycin-resistant; CSES, clindamycin-susceptible erythromycin-susceptible; CSEI, clindamycin-susceptible erythromycin-intermediate; CREI, clindamycin-resistant erythromycin-intermediate; CIES, clindamycin-intermediate erythromycin-susceptible; OAT, Orthologous Average Nucleotide Identity Tool.
|
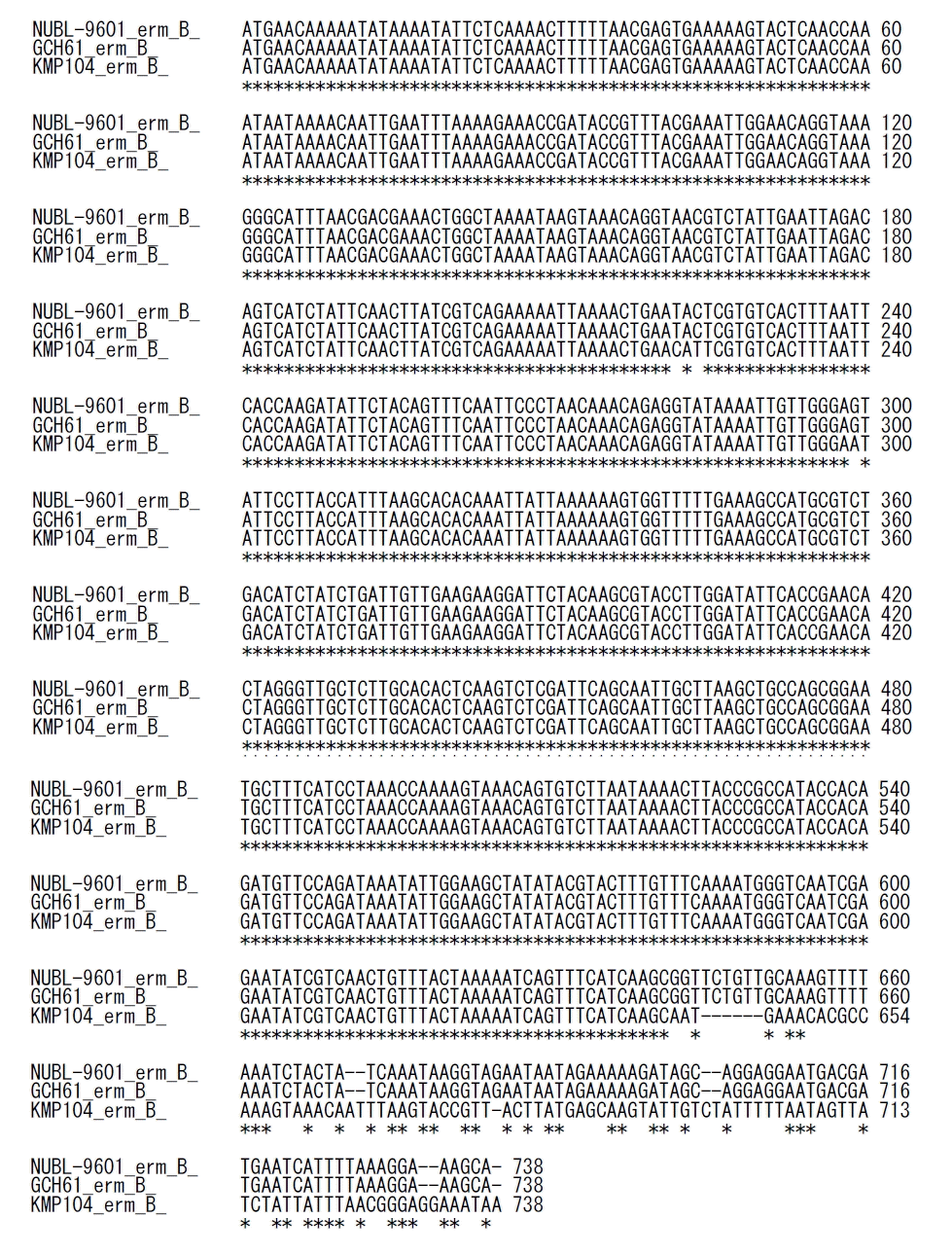 | Fig. 3Comparison of sequences of GCH61, NUBL-9601, and KMP104. Sequence of erm(B) from isolate GCH61 (accession number VYRN00000000) with the CRES phenotype contained C222T (N74N), T224C (I75T), and A299G (N100S) nucleotide (amino acid) substitutions, in addition to the insertion of an IS1216E element at nucleotide position 642, which resulted in the deletion of a segment spanning nucleotides 642–738 (97 bp). This sequence was identical to that from the CRES isolate NUBL-9601 (accession number LC430933). The sequence of erm(B) from the isolate KMP104 was used as a reference (RefSeq accession number DQ355148).
Abbreviation: CRES, clindamycin-resistant erythromycin-susceptible.
|




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download