The ongoing coronavirus disease 2019 (COVID-19) pandemic began in December 2019 in Wuhan, China, with more than 13,824,739 confirmed cases as of July 18th, 2020 [
1]. In Korea, the first COVID-19 case was confirmed on January 20th, 2020. Rapid viral transmission during the early phase of COVID-19 has highlighted the importance of laboratory diagnosis [
2,
3]. Molecular testing is the reference standard for COVID-19 diagnosis, and real-time reverse transcription-PCR (rRT-PCR) is the preferred one [
2,
4,
5]. The ability of clinical laboratories to perform an appropriate molecular testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection is critical for an effective response to the ongoing pandemic. Rapid response system, including an emergency use authorization (EUA) system for emerging infectious diseases, were established by the Korean Centers for Disease Control and Prevention (KCDC), Korean Society for Laboratory Medicine (KSLM), and the Korean Association of External Quality Assessment Service [
6,
7]. The first commercial rRT-PCR assay for detecting SARSCoV-2 was granted EUA by the Ministry of Food and Drug Safety in Korea on February 4th, 2020, and SARS-CoV-2 rRT-PCR testing by clinical laboratories began by February 7th, 2020. However, as the clinical performance of this assay reagent had not been fully evaluated, stringent and continuous quality control was required [
7-
9]. Therefore, KSLM COVID-19 Task Force established an online laboratory surveillance system to monitor SARS-CoV-2 rRT-PCR testing capacities and results. We analyzed the surveillance data obtained using this system from February 7th to June 4th, 2020.
SARS-CoV-2 rRT-PCR data were collected from 97 clinical laboratories, including 84 medical institutions and 13 independent clinical laboratories; each laboratory submitted the data to KSLM daily beginning from February 7th, 2020. Public health laboratories, such as the KCDC and local government laboratories, were excluded from this study. Submitted data included: (1) the number of tests performed, including the number that gave positive, negative, and invalid/indeterminate results for SARS-CoV-2, with invalid denoting “failure of both internal control and target gene amplification” and indeterminate denoting that “only some genes were amplified”; (2) detailed information on each positive result, including sample type, assay reagent and equipment used, and threshold cycle (Ct) values for each target gene; and (3) parallel test results to confirm the acceptability of new reagent lot. We collected and reviewed the submitted data once daily and checked errors to ensure data quality. The Institutional Review Board (IRB) of the National Health Insurance Medical Center, Goyang, Korea (IRB No. NHIMC 2020-04-025) approved the study.
We analyzed Spearman correlation to examine the associations between the Ct values of genes. We used the Wilcoxon signed-rank test to compare the Ct values of paired upper respiratory tract (URT) and lower respiratory tract (LRT) samples with Bonferroni correction for multiple comparisons. P < 0.05 was considered significant. Statistical analyses were performed using the MedCalc Statistical Software version 19.2.1 (MedCalc Software Ltd, Ostend, Belgium).
In total, 1,890,319 SARS-CoV-2 rRT-PCR testing was performed, 2.3% of which were positive (
Table 1). The testing number and participating laboratories significantly increased by the third week of the study period (February 7th to June 4th, 2020), proportionate to the increase in confirmed cases; with over 70,000 testing being performed weekly after the initial two weeks, the daily testing capacity (34,193 tests) peaked on May 29th, 2020 (
Fig. 1).
Fig. 1
Monitoring of SARS-CoV-2 rRT-PCR testing performed by clinical laboratories in Korea from February 7th to June 4th, 2020. Confirmed case numbers were derived from the KCDC data (
http://ncov.mohw.go.kr/).
Abbreviations: COVID-19, coronavirus disease 2019; EUA, emergency use authorization; KCDC, Korean Centers for Disease Control and Prevention; MFDS, Ministry of Food and Drug Safety; rRT-PCR, real-time reverse transcription PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
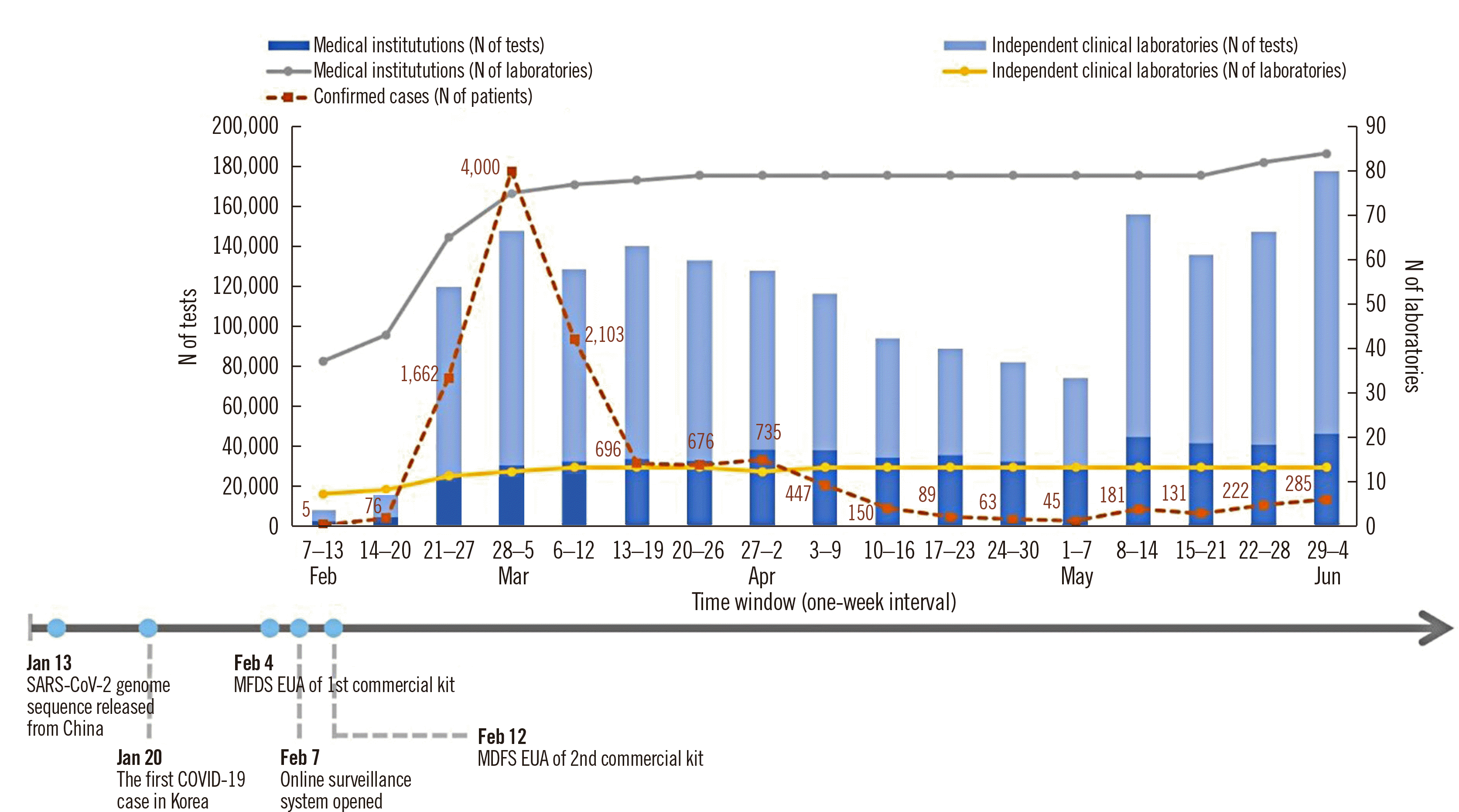

Table 1
Number of SARS-CoV-2 rRT-PCR testing performed by clinical laboratories in Korea from February 7th to June 4th, 2020
|
Medical institutions, N (%) |
Independent clinical laboratories, N (%) |
Total, N (%) |
|
Tests |
533,615 (100) |
1,356,704 (100) |
1,890,319 (100) |
|
Positive results |
13,772 (2.6) |
29,797 (2.2) |
43,569 (2.3) |
|
Negative results |
518,023 (97.1) |
1,319,162 (97.2) |
1,837,185 (97.2) |
|
Invalid/indeterminate results |
1,820 (0.3) |
7,745 (0.6) |
9,565 (0.5) |

Of the positive results, 13,006 results were initially obtained from newly diagnosed COVID-19 patients. Majority of the tests were conducted using the PowerChek 2019-nCoV Real-time PCR Kit (Kogenebiotech, Seoul, Korea), targeting the envelope (
E) and RNA-dependent RNA polymerase (
RdRp) genes, and the Allplex 2019-nCoV Assay (Seegene, Seoul, Korea), targeting the
E, RdRp, and nucleocapsid (
N) genes; these were the first and second commercial assays to receive EUA, respectively (
Supplemental Data Table S1). The parallel test results showed low lot‐to‐lot reagent variation (
Supplemental Data Table S2). Strong correlations were observed between the
E and
RdRp/
N Ct values using PowerChek and Allplex (Spearman’s rho > 0.98,
P < 0.001), and lower Ct values were obtained for
E than for the
RdRp or
N genes. The median Ct value difference between
E and
RdRp was 0.40 (95% confidence interval [CI], 0.38–0.43) for PowerChek. For Allplex, the median Ct value differences between genes were 1.25 for
E and
RdRp (95% CI, 1.24–1.27) and 2.42 for
E and
N (95% CI, 2.40–2.44).
Of the initial positive results, results from paired URT and LRT samples were used to evaluate positive result characteristics according to the sample types. Results for 3,832 samples were obtained from 1,916 paired URT (nasopharyngeal swab [NPS] or nasopharyngeal/oropharyngeal swab [NPS/OPS]) and LRT (sputum) samples, including 909 sets of NPS and sputum samples and 1,007 sets of NPS/OPS and sputum samples. The distributions of
E,
RdRp, and
N gene Ct values in the paired samples are presented in
Table 2. No statistically significant differences in gene Ct values were observed between the paired URT and LRT samples, except in the
N gene for NPS and sputum samples.
Table 2
Comparison of Ct values in paired upper and lower respiratory tract samples obtained from newly diagnosed COVID-19 patients (N =1,916)
|
SARS-CoV-2 target gene |
NPS vs. sputum (N=909) |
NPS/OPS vs. sputum (N=1,007) |
|
|
|
Ct value, Median (IQR) |
Ct difference, Median (IQR) |
Ct value, Median (IQR) |
Ct difference, Median (IQR) |
|
|
|
|
|
NPS |
Sputum |
Sputum-NPS |
P*
|
NPS/OPS |
Sputum |
Sputum-NPS |
P*
|
|
E
|
24.61 (19.20-29.51) |
24.57 (19.57-29.06) |
0.22 (-0.16-0.59) |
>0.05 |
24.69 (18.59-29.65) |
24.82 (19.24-29.52) |
0.19 (-0.14-0.51) |
>0.05 |
|
RdRp
|
25.65 (20.33-30.42) |
25.82 (20.95-30.12) |
0.28 (-0.07-0.64) |
>0.05 |
25.66 (19.97-30.86) |
25.89 (20.49-30.72) |
0.29 (-0.03-0.61) |
>0.05 |
|
N
|
26.97 (21.58-31.29) |
27.12 (22.31-31.34) |
0.49 (0.11-0.88) |
0.034 |
26.89 (21.22-31.79) |
27.18 (21.97-31.95) |
0.43 (0.07-0.80) |
>0.05 |

Surveillance for emerging pathogens is a common practice; however, daily surveillance is uncommon [
10,
11]. The laboratory-based SARS-CoV-2 surveillance system in our study can provide daily laboratory data for monitoring SARS-CoV-2 rRTPCR testing results in Korea and is being used successfully in a timely fashion. Our study showed that clinical laboratories in Korea responded quickly to the COVID-19 outbreak, with SARSCoV-2 rRT-PCR testing beginning 16 days after the first confirmed case in the country, and that the testing capacity increased rapidly thereafter. Of note, clinical laboratories consistently conducted over 70,000 tests weekly during the study period, although the number of confirmed patients sharply decreased since the peak in the first week of March 2020.
Recent studies have reported that SARS-CoV-2 detection sensitivity is greater for LRT samples than for URT samples [
12-
15]. Using paired sample analysis, Lin,
et al. [
14] demonstrated that the detection rates of SARS-CoV-2 for sputum samples were significantly higher than those for throat swabs. In our study, paired sample analysis, with positive results from both URT and LRT samples, showed that the Ct value differences between sample types were generally not significant. Although the
N gene Ct value differences between NPS and sputum were significant, the median Ct difference (0.49) was low. As these results were derived from initial positive results from newly diagnosed COVID-19 patients, it is possible that several patients were paucisymptomatic or asymptomatic, without productive sputum.
This study has several limitations. We have reported not the number of COVID-19 patients but the number of SARS-CoV-2 rRT-PCR testing. Additionally, we did not collect clinical information, as the study was based on laboratory surveillance data regarding testing capacities and positive result details. In addition, detailed information, such as sample type, was not available for all negative results.
In summary, this laboratory surveillance report for SARSCoV-2 rRT-PCR testing provided a timely assessment of testing capacities of clinical laboratories and positive result characteristics, according to sample type and assays, for the first few months of the COVID-19 outbreak in Korea. Clinical laboratories rapidly expanded their testing capacity in response to the COVID-19 outbreak, with a peak daily testing capacity of 34,193 tests. This rapid expansion in testing capacity is a critical component of the national response to the ongoing pandemic.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download