INTRODUCTION
The clinical significance of HLA-DQ antibodies in kidney transplantation (KT) has been highlighted by the development of molecular HLA typing and single-antigen bead (SAB) assay. It is known that 15%–30% of patients develop donor-specific antibodies (DSA) within three years of KT, and 33%–77% of them develop DQ DSA, which is associated with poor graft outcome [
1–
7]. Given its clinical significance, HLA-DQ typing for deceased donors is required by Eurotransplant (Austria, Belgium, Croatia, Germany, Hungary, Luxembourg, the Netherlands, and Slovenia) and the United Network for Organ Sharing (USA) [
8,
9].
However, HLA-DQ typing for deceased donors is not yet required in Korea. Furthermore, the Korean Network for Organ Sharing has no regulations regarding deceased donor specimen storage for additional HLA typing, making retrospective HLA typing nearly impossible. Therefore, when a patient develops DQ antibodies after deceased donor KT, it is impossible to determine whether these antibodies are donor-specific or not.
HLA shows typical haplotype characteristics according to individual ethnic groups [
10,
11]. Therefore, HLA-DQ can be predicted using HLA haplotype frequency and linkage disequilibrium (LD) data. We developed a DQ prediction program based on HLA-A, -B, and -DR and analyzed the clinical significance of the predicted DQ (pDQ) in KT recipients. To the best of our knowledge, this is the first study to predict HLA-DQ using an artificial neural network (ANN) and to evaluate its clinical significance.
Go to :

MATERIALS AND METHODS
Data source and study population
HLA typing data, SAB assay data, and patients’ medical records were retrospectively reviewed. To develop the DQ prediction program and evaluate its accuracy, low-resolution HLA-A, -B, -C, -DR, -DQ typing data of 6,006 Korean patients registered at Asan Medical Center, Seoul, Korea from October 2005 to April 2019 were used. The data of 5,603 patients were analyzed in terms of allele and haplotype frequencies and used to train an ANN. The data of the remaining 403 patients were used to evaluate the accuracy of the HLA-DQ prediction program (
Fig. 1A).
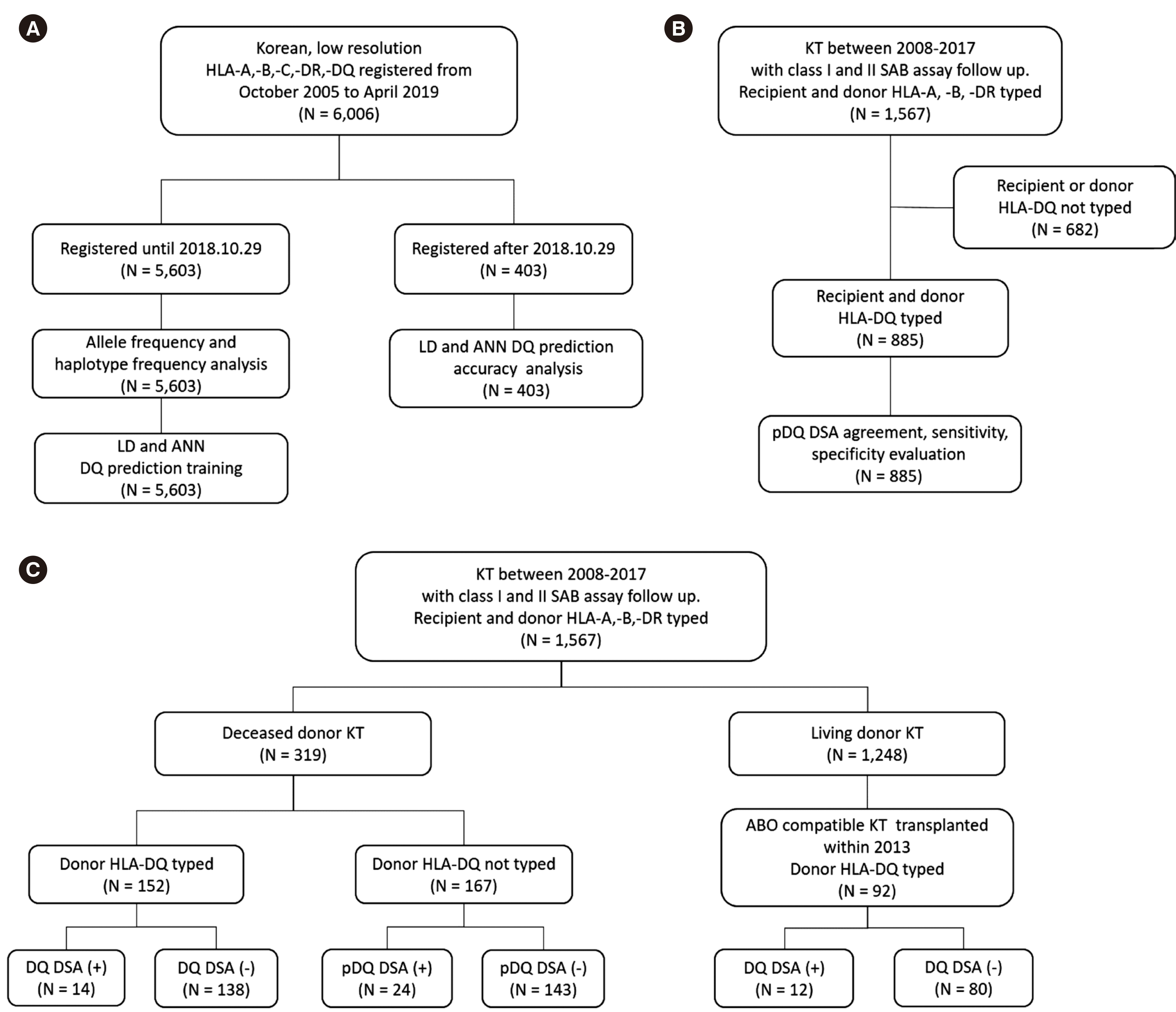 | Fig. 1
Patient and data selection flowchart. (A) HLA-DQ prediction program development and evaluation. (B) Evaluation of pDQ DSA agreement, sensitivity, and specificity. (C) Evaluation of DQ and pDQ DSA clinical significance.
Abbreviations: ANN, artificial neural network; DSA, donor specific antibody; KT, kidney transplantation; LD, linkage disequilibrium; pDQ, predicted DQ; SAB, single-antigen bead.

|
To evaluate pDQ DSA agreement, sensitivity, specificity, and clinical significance, we enrolled Korean patients who underwent KT between January 2008 and December 2017, whose recipient/donor HLA-A, -B, -DR typing data were available, and who had undergone follow-up class I and II SAB assays. Patients who underwent kidney re-transplantation or any other type of transplantation were excluded. A total of 1,567 patients met the inclusion criteria. For pDQ DSA agreement, sensitivity, and specificity analyses, recipients/donors with unknown HLA-DQ were excluded. In total, 885 recipients/donors with available HLA-DQ typing data were included (
Fig. 1B). To evaluate the clinical significance of pDQ DSA, previously selected 1,567 patients were divided into the deceased donor KT and living donor KT groups. The deceased donor KT group (N = 319) was subdivided according to donor HLA-DQ availability and DQ or pDQ DSA positivity. In the living donor KT group (N = 1,248), 92 patients who had ABO-compatible KT in 2013 were selected and subdivided according to DQ DSA positivity (
Fig. 1C). This study was approved by the Institutional Review Board of Asan Medical Center (S2019-0353-0001).
HLA typing and DSA detection
The HLA-A, -B, -C of KT recipients and living donors were analyzed using AVITA Cross sequence-based typing (SBT) (Biowithus, Seoul, Korea), while their HLA-DR and -DQ were analyzed using AVITA SBT (Biowithus). HLA-A, -B, -C, -DR of deceased donors were typed using a PCR/sequence-specific primer (SSP) kit (BioSewoom Inc., Seoul, Korea). Since November 2015, deceased donor HLA-DQ has been typed using AVITA SBT (Biowithus). HLA DSA was screened using LABScreen Single Antigen assay (One Lambda, Canoga Park, CA, USA). Normalized mean fluorescence intensity (MFI) was calculated using an HLA fusion software (One Lambda), and an MFI of 1,000 was set as a cutoff.
Development of HLA-DQ prediction programs
Two DQ prediction programs were developed, one based on LD and the other on a multilayer perceptron ANN.
The LD algorithm used Lewontin’s
D´ to predict HLA-DQ using HLA-B and -DR information [
12]. In each case, possible HLAB-DR haplotypes were generated, and the frequency of each haplotype was calculated. The
D´ was then calculated between possible HLA-B-DR haplotypes and every DQ type in our database. The HLA-DQ showing the highest
D´, and the most frequent HLA-B-DR-DQ haplotype was chosen as the pDQ. The second most likely pDQ, showing the second-highest D’ or the second most frequent HLA-B-DR-DQ haplotype, was also generated by the LD algorithm. The second most likely pDQ was only applied for pDQ accuracy evaluation.
The ANN model predicted the HLA-DQ with HLA-A, -B, and -DR information using feed-forward neural network. In the input layer, the categorical data allele was encoded into a sparse vector, a one hot vector. A total of six one hot vectors are converted for each sample to represent HLA-A, -B, and -DR alleles. The embedding layer multiplied the one hot sparse vector with the embedding matrix to convert it into dense vectors. Hidden layers consisted of multiple layers, and performed linear transformation and activation functions repeatedly. Finally, the output layer consisted of a classifier model with two heads. In each head, the input vector was converted to a one hot vector through a linear transformation and a soft-max function, and converted to a categorical value, DQ alleles.
pDQ accuracy and pDQ DSA agreement, sensitivity, and specificity
pDQ genotype accuracy and allele accuracy were evaluated by comparing the pDQ and typed DQ data. DSA was reanalyzed with recipient/donor pDQ instead of typed DQ from preexisting SAB assay data. Any DQ DSA detected through the reanalysis of the SAB assay using donor and recipient pDQ data was labeled pDQ DSA.
pDQ DSA agreement was determined by comparing the presence of DQ DSA and pDQ DSA in the 1,970 SAB assay results from 885 recipients. If pDQ DSA was detected in DQ DSA-positive case or not detected in DQ DSA-negative case, it was considered as an agreement. The sensitivity (number of SAB assay results with pDQ DSA among DQ DSA-positive ones), specificity (number of SAB assay results without pDQ DSA among DQ DSA-negative ones), false-positive, and false-negative rates of pDQ DSA were estimated using the presence of the SAB assay DQ DSA as a standard.
Clinical significance of pDQ DSA
The post-transplantation outcomes were classified into five groups: biopsy-proven antibody-mediated rejection (ABMR) [
13], biopsy-suspicious ABMR, clinically suspicious ABMR, biopsy-proven non-ABMR, and clinically non-ABMR. pDQ DSA was also regarded as serological evidence of DSA.
Statistical analysis
The ALLELE and HAPLOTYPE procedures of SAS (version 9.4; SAS Institute Inc., Cary, NC, USA) were used to analyze the allele and haplotype frequencies, respectively. Differences in clinical course among the patient groups were analyzed using the chi-square test. Two-sided P < 0.05 was considered statistically significant. Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA) and SPSS Statistics for Windows version 19.0 (IBM Corp., Armonk, NY, USA) were also used for the analyses.
Go to :

DISCUSSION
It was thought that if the HLA-DR was compatible serologically, HLA-DQ would not affect KT outcomes, as HLA-DR and HLA-DQ show strong LD [
14,
15]. However, molecular typing has revealed that HLA-DQ is discordant in 15%–26% of HLA-DR-matched patients [
16,
17].
We developed two different DQ prediction programs using the LD algorithm and an ANN. The genotype accuracies of LD pDQ and ANN pDQ were over 75%. When the second most likely pDQ was included, the LD pDQ genotype accuracy increased to 92.6%. The accuracy was poorest for DQ8. This is partly due to the LD distribution of the HLA-B-DR-DQ haplotype with which DQ8 is associated. We evaluated the LD between HLA-B-DR and DQ of 74 HLA-B-DR-DQ8 haplotypes in Korean patients. Among the HLA-B-DR haplotypes associated with DQ8, 61 (82.4%) had the same or higher D´ with DQ, except for DQ8 (data not shown).
The SAB assay results were reanalyzed using pDQ. Both LD and ANN pDQ DSA showed agreement, sensitivity, and specificity over 95%. The MFI of all false-positive pDQ DSAs was < 5,000. Previous studies have shown that patients with allograft loss had a significantly higher DQ DSA MFI (6,000–16,000) than those without allograft loss [
3,
18]. Thus, antibodies with an MFI < 5,000 detected as false positives by the DQ prediction program were considered to be of low clinical significance.
In this study, ABMR incidence was significantly higher in DQ or pDQ DSA-positive patients than in DQ or pDQ DSA-negative patients (64.0% vs. 7.8%,
P < 0.001). ABMR incidence was similar to that in the study by DeVos,
et al. [
2], wherein ABMR incidence in KT recipients without DSA, recipients with DQ-only DSA, and recipients with DQ + other DSA was 11% (31/285), 21% (7/33), and 67% (10/15), respectively. No significant difference in ABMR incidence was observed between the DQ DSA- and pDQ DSA-positive groups and between the DQ DSAand pDQ DSA-negative groups. These results suggest that DQ DSA and pDQ DSA have similar clinical characteristics.
Of the patients who underwent a deceased donor KT with unknown donor DQ, patient 1 had pDQ9 DSA MFI increased from 9,315 to 15,205 during the follow-up period, with worsening glomerulopathy and peritubular capillaritis (
Table 5). The clinical course of the patient and the accuracy of the DQ prediction program suggest that pDQ9 is likely an actual DSA. Although the pDQ9 DSA MFI exceeded 10,000 in 2016 and 2017, the patient did not receive ABMR treatment at that time. Patient 4 had normal but increasing creatinine levels at three yrs posttransplantation (
Table 5). DQ7 antibody was identified but not reported as a DSA, as the donor DQ was not available. The patient received only steroid treatment, and his creatinine level remained normal. In this case, it might have been helpful to follow up the SAB assay and review the MFI trends of the DQ7 antibody.
Patients 2, 3, 4, and 5 were C4d-negative through kidney biopsy. ABMR can be diagnosed based on kidney biopsy results and the presence of DSA; however, when DSA results are not available, the C4d status can serve as an alternative for ABMR diagnosis [
13]. Unfortunately, C4d has low sensitivity and, even among microvascular injury and DSA, 20% of the patients are C4d-negative [
19]. Therefore, the revised Banff 2017 strongly recommends DSA analysis [
13]. pDQ DSA analysis would be particularly useful for patients with unclear biopsy results, such as those who are C4d-negative, in the context of donors who have not been HLA-DQ typed.
Patient 6 had a pDQ DSA with an MFI persistently exceeding 10,000. This patient had an elevated creatinine level and underwent steroid treatment. As kidney biopsy was not available for this patient, it was difficult to determine whether the pDQ DSA was a true DSA. Given that the pDQ DSA MFI exceeded 10,000 and increased during the follow-up period, additional assays might be needed to diagnose ABMR.
Taken together, our results suggest that if DQ antibodies are detected in the SAB assay of a patient with an unknown donor DQ, the DQ prediction program can help diagnose and treat ABMR. If the patient presents pDQ DSA, we recommend performing a SAB assay periodically and follow up pDQ DSA MFI, in addition to tests such as kidney scan and biopsy. More active treatment should be considered, if the pDQ DSA MFI exceeds 5,000 or other DSA is also present. In addition, even if pDQ DSA is not detected, if the same DQ antibody is repeatedly identified by the SAB assay, the possibility of an incorrect DQ prediction should be considered. In such cases, the SAB assay can be performed using the second most likely pDQ according to the LD algorithm.
Our study had several limitations. First, pDQ DSA agreement does not indicate an actual match between pDQ DSA and DQ DSA, but a comparison of the presence of a pDQ and DQ DSA. To assess the clinical significance of DQ or pDQ DSA, the ABMR treatment frequency was analyzed. Long-term observation of cases for ABMR-free survival or graft survival rates would better reflect the clinical course. Lastly, usage of the HLA-DQ prediction programs is limited to our institution.
In conclusion, we developed HLA-DQ prediction programs using the LD algorithm and ANN. Although experts agree that HLA-DQ typing is necessary for deceased donors, cost remains an issue, and it will take time for existing policies to address this issue. As there were some inaccuracies in the prediction programs developed in this study, a policy of HLA-DQ typing for deceased donors should be established as promptly as possible in Korea.
Go to :






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download