INTRODUCTION
MATERIALS AND METHODS
Patient samples
Rotavirus G and P genotyping
Lewis antigen phenotyping
PCR and sequencing of the secretor and Lewis genes (FUT2 and FUT3)
Analyses of amino acid sequences and three-dimensional (3D) structure of VP8*
Statistical analysis
RESULTS
Rotavirus genotype, erythrocyte Lewis antigen phenotypes, and FUT2/FUT3 gene variants in rotavirus infected patients
Table 1
| Rotavirus genotype | Case number | Sex/Age | Erythrocyte Lewis phenotype |
Minor allele homozygosity (FUT2 c.418, c.461, c.772) |
Minor allele homozygosity (FUT3 c.59, c.202, c.314, c.508, or c.1067) |
|---|---|---|---|---|---|
| G8P[6] |
1 19RP-09 |
M/15 days | Lea(3+) Leb(-)* | c.418A > T | - |
|
2 19RP-11 |
M/8 days | Lea(1+) Leb(2+) | - | - | |
|
3 19RP-26 |
M/20 days | Lea(−) Leb(−) | - | c.508G > A | |
|
4 19RP-35 |
M/15 days | Lea(−) Leb(−) | c.418A > T | c.59T > G | |
|
5 19RP-38 |
M/9 days | Lea(−) Leb(−) | - | c.59T > G, c.508G > A | |
| G1P[8] |
6 19RP-23 |
F/3 yr | Lea(−) Leb(4+) | - | - |
| G8P[8] |
7 19RP-04 |
M/6 yr | Lea(−) Leb(4+) | - | - |
|
8 19RP-07 |
M/4 yr | Lea(−) Leb(4+) | - | - | |
|
9 19RP-31 |
F/4 yr | Lea(−) Leb(4+) | - | - | |
| G9P[8] |
10 19RP-02 |
F/3 yr | Lea(−) Leb(4+) | - | - |
|
11 19RP-15 |
F/6 yr | Lea(−) Leb(3+) | - | - | |
|
12 19RP-24 |
F/ 20 days | Lea(3+) Leb(3+) | - | - | |
| G2P[4] |
13 19RP-13 |
M/5 yr | Lea(−) Leb(4+) | - | - |
|
14 19RP-17 |
F/10 yr | Lea(−) Leb(3+) | - | - | |
|
15 19RP-21 |
F/1 yr | Lea(−) Leb(4+) | - | - | |
|
16 19RP-32 |
M/9 yr | Lea(−) Leb(3+) | - | - |
Amino acid sequence of the VP8* portion of the VP4 capsid protein
Table 3
| Amino acid | 81 | 91 | 101 | 111 | 121 | 130 | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wa_G1P[8]_JX406750* | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | V | A | I | E | P | H | V | N | P | V | D | R | Q | Y | T | I | F | G | E | S | K | Q | F | N | V |
| 19RP-23_G1P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-04_G8P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-07_G8P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-31_G8P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-02_G9P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-15_G9P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| 19RP-24_G9P[8] | W | I | L | I | N | S | N | T | N | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | D | P | V | D | R | Q | Y | N | V | F | G | E | N | K | Q | F | N | V |
| DS-1_G2P[4]_HQ650119* | W | L | L | I | S | S | N | T | N | G | V | V | Y | E | S | T | N | N | N | D | F | W | T | A | V | I | A | V | E | P | H | V | S | Q | T | N | R | Q | Y | I | L | F | G | E | N | K | Q | F | N | V |
| 19RP-13_G2P[4] | W | L | L | I | S | S | N | T | D | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | S | P | T | N | R | Q | Y | V | L | F | G | E | N | K | Q | F | N | V |
| 19RP-17_G2P[4] | W | L | L | I | S | S | N | T | D | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | S | P | T | N | R | Q | Y | V | L | F | G | E | N | K | Q | F | N | V |
| 19RP-21_G2P[4] | W | L | L | I | S | S | N | T | D | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | S | Q | T | N | R | Q | Y | V | L | F | G | E | N | K | Q | F | N | I |
| 19RP-32_G2P[4] | W | L | L | I | S | S | N | T | D | G | V | V | Y | E | S | T | N | N | S | D | F | W | T | A | V | I | A | V | E | P | H | V | S | P | T | N | R | Q | Y | V | L | F | G | E | N | K | Q | F | N | V |
| RV3_P[6]_U16299* | W | I | L | L | N | P | T | N | Q | Q | V | V | L | E | G | T | N | K | T | D | I | W | V | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | T | K | Q | I | T | V |
| 19RP-09_G8P[6] | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| 19RP-26_G8P[6] | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| 19RP-35_G8P[6] | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| 19RP-38_G8P[6] | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| RN-001_KOR_G4P[6]_MK953605 | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| 27_ 2017_KOR_G8P[6] [8] | W | I | L | L | N | P | T | D | Q | Q | V | V | L | E | S | T | N | K | T | D | I | W | I | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | M | R | Q | I | T | I |
| P[6] RV3_U16299* | W | I | L | L | N | P | T | N | Q | Q | V | V | L | E | G | T | N | R | T | D | I | W | V | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | T | K | Q | I | T | V |
| P[6] Nicaragua_JN129097* | W | I | L | L | N | P | T | N | Q | A | V | V | L | E | G | T | N | R | T | D | V | W | V | A | L | L | L | I | E | P | N | T | P | S | Q | S | R | Q | Y | T | L | F | G | E | I | K | Q | I | S | V |
| P[6] Vietnam 2007_LC095922* | W | I | L | L | S | P | T | N | Q | Q | V | V | L | E | G | T | N | R | T | D | V | W | V | A | L | L | L | I | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | T | K | Q | I | T | V |
| P[6] Hungary 2002_KF835915* | W | I | L | L | N | P | T | N | Q | K | V | V | L | E | G | T | N | R | A | D | V | W | V | A | I | L | L | I | E | P | N | T | I | N | Q | S | R | Q | Y | T | L | F | G | E | T | K | Q | I | T | I |
| P[6] China 2013_KF726067* | W | I | L | L | S | P | T | N | Q | Q | V | V | L | E | G | T | N | R | T | D | V | W | V | A | L | L | L | V | E | P | N | V | T | N | Q | S | R | Q | Y | T | L | F | G | E | T | K | NI | I | T | V |
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Amino acid | 131 | 141 | 151 | 161 | 171 | 180 | ||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Wa_G1P[8]_JX406750* | S | N | D | - | - | I | W | K | F | L | E | M | F | R | S | S | S | Q | N | E | F | Y | N | R | R | T | L | T | S | D | T | R | L | V | G | I | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| 19RP-23_G1P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | S | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-04_G8P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | G | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-07_G8P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | G | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-31_G8P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | S | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-02_G9P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | S | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-15_G9P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | S | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| 19RP-24_G9P[8] | R | N | D | - | - | I | W | K | F | L | E | M | F | R | G | S | S | Q | S | D | F | Y | N | R | R | T | L | T | S | D | T | K | L | V | G | I | L | K | Y | G | G | R | I | W | T | F | H | G | E | T |
| DS-1_G2P[4]_HQ650119* | E | N | N | - | - | I | W | K | F | F | E | M | F | K | G | S | S | Q | G | D | F | S | N | R | R | T | L | T | S | S | N | R | L | V | G | M | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| 19RP-13_G2P[4] | E | N | S | - | - | I | W | K | F | F | E | M | F | K | G | S | S | Q | S | D | F | S | N | R | R | T | L | T | S | D | N | R | L | V | G | M | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| 19RP-17_G2P[4] | E | N | S | - | - | I | W | K | F | F | E | M | F | K | G | S | S | Q | S | D | F | S | N | R | R | T | L | T | S | D | N | R | L | V | G | M | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| 19RP-21_G2P[4] | E | N | S | - | - | T | W | K | F | F | E | M | F | K | G | S | S | Q | S | D | F | S | N | R | R | T | L | T | S | N | N | R | L | V | G | M | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| 19RP-32_G2P[4] | E | N | S | - | - | I | W | K | F | F | E | M | F | K | G | S | S | Q | S | D | F | S | N | R | R | T | L | T | S | D | N | R | L | V | G | M | L | K | Y | G | G | R | V | W | T | F | H | G | E | T |
| RV3_P[6]_U16299* | E | N | N | T | N | K | W | K | F | F | E | M | F | R | S | S | V | S | A | E | F | Q | H | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | Y | N | S | V | W | T | F | H | G | E | T |
| 19RP-09_G8P[6] | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| 19RP-26_G8P[6] | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| 19RP-35_G8P[6] | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| 19RP-38_G8P[6] | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| RN-001_KOR_G4P[6]_MK953605 | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| 27_ 2017_KOR_G8P[6] [8] | E | N | N | - | - | T | W | K | F | F | E | V | F | R | R | T | A | G | A | E | F | Q | H | K | H | T | L | T | S | D | T | K | L | A | G | F | L | K | L | Y | N | S | V | W | T | F | Y | G | E | T |
| P[6] RV3_U16299* | E | N | N | T | N | K | W | K | F | F | E | M | F | R | S | S | V | S | A | E | F | Q | H | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | Y | N | S | V | W | T | F | H | G | E | T |
| P[6] Nicaragua_JN129097* | E | N | N | T | N | K | W | K | F | F | E | M | F | R | N | N | I | N | S | E | F | Q | H | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | G | G | R | V | W | T | F | H | G | E | T |
| P[6] Vietnam 2007_LC095922* | E | N | N | T | N | E | W | K | F | Y | E | M | F | R | N | S | A | N | V | E | F | Q | H | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | G | G | R | V | W | T | F | H | G | E | T |
| P[6] Hungary 2002_KF835915* | E | N | N | S | S | K | W | K | F | F | E | M | F | R | N | N | A | S | A | E | F | Q | H | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | G | G | R | V | W | T | F | H | G | E | T |
| P[6] China 2013_KF726067* | E | N | N | T | N | K | W | K | F | Y | E | M | F | R | N | N | A | S | A | E | F | Q | P | K | R | T | L | T | S | D | T | K | L | A | G | F | L | K | H | G | G | R | V | W | T | F | H | G | E | T |
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Amino acid | 181 | 191 | 201 | 211 | 221 | 229 | ||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Wa_G1P[8]_JX406750* | P | R | A | T | T | D | S | S | S | T | A | N | L | N | N | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-23_G1P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-04_G8P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-07_G8P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-31_G8P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | D | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-02_G9P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-15_G9P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-24_G9P[8] | P | R | A | T | T | D | S | S | N | T | A | N | L | N | G | I | S | I | T | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| DS-1_G2P[4]_HQ650119* | P | R | A | T | T | D | S | S | N | T | A | D | L | N | N | I | S | I | I | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-13_G2P[4] | P | R | A | T | T | D | S | S | S | T | A | D | L | N | N | I | S | I | I | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-17_G2P[4] | P | R | A | T | T | D | S | S | S | T | A | D | L | N | N | I | S | I | I | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-21_G2P[4] | P | R | A | T | T | D | S | S | N | T | A | D | L | N | N | I | S | I | I | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-32_G2P[4] | P | R | A | T | T | D | S | S | S | T | A | D | L | N | N | I | S | I | I | I | H | S | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| RV3_P[6]_U16299* | P | H | A | T | T | D | Y | S | S | T | S | N | L | S | E | V | E | T | T | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | T | G | L | |||||||
| 19RP-09_G8P[6] | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | N | G | L | P | P | I | Q | N | T | |
| 19RP-26_G8P[6] | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | D | Y | I | N | N | G | L | P | P | M | Q | N | T | |
| 19RP-35_G8P[6] | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | L | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | N | G | L | P | P | M | Q | N | T | |
| 19RP-38_G8P[6] | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | L | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | N | G | L | P | P | M | Q | N | T | |
| RN-001_KOR_G4P[6]_MK953605 | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | T | G | L | P | P | M | Q | N | T | |
| 27_2017_KOR_G8P[6] [8] | P | H | A | T | T | D | Y | L | L | T | S | N | L | S | E | V | E | A | V | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | N | G | L | P | P | M | Q | N | T | |
| P[6] RV3_U16299* | P | H | A | T | T | D | Y | S | S | T | S | N | L | S | E | V | E | T | T | I | H | V | E | F | Y | I | I | P | R | S | Q | E | S | K | C | V | E | Y | I | N | T | G | L | |||||||
| P[6] Nicaragua_JN129097* | P | H | A | T | T | D | Y | S | T | S | N | L | S | E | V | I | E | T | V | I | H | A | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | T | G | L | |||||||
| P[6] Vietnam 2007_LC095922* | P | H | A | T | T | D | Y | S | S | S | N | L | S | E | V | I | E | T | V | I | H | T | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | T | G | L | |||||||
| P[6] Hungary 2002_KF835915* | P | N | A | T | T | D | Y | S | T | S | N | L | S | P | D | I | E | T | V | I | H | T | D | F | Y | I | I | P | R | S | Q | E | S | K | C | K | E | Y | I | N | T | G | L | |||||||
| P[6] China 2013_KF726067* | P | H | A | T | T | D | Y | S | S | S | N | L | S | E | V | V | E | T | V | I | H | T | E | F | Y | I | I | P | R | S | Q | E | S | K | C | N | E | Y | I | N | T | G | L | |||||||




 PDF
PDF Citation
Citation Print
Print



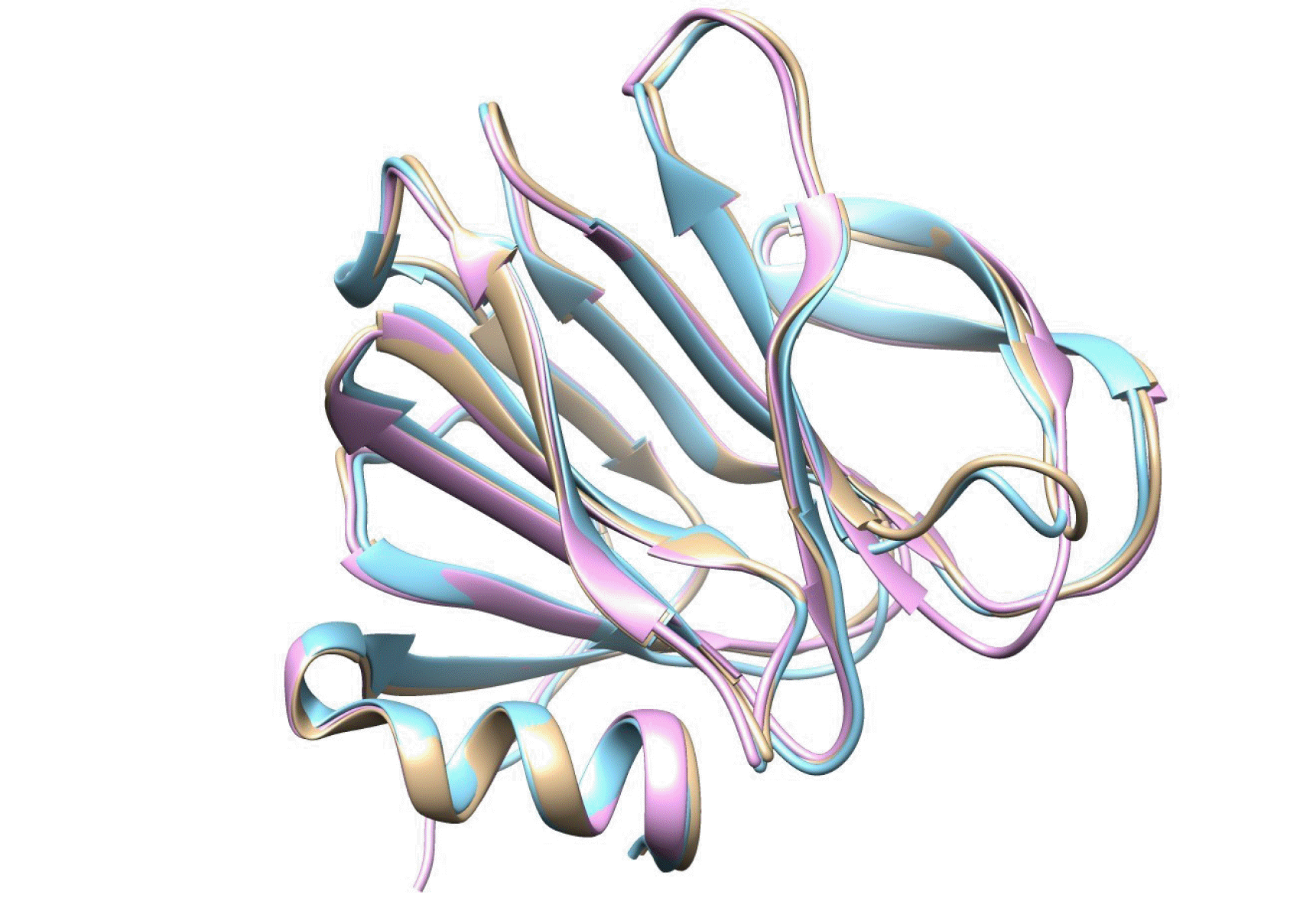
 XML Download
XML Download