Abstract
Peptides are short chain of amino acids linked by peptide bonds. They are widely used as effective and biocompatible active ingredients in cosmetic industry. In this study, we developed novel peptide mixture and identified its anti-pigmentation effect on melanocytes and keratinocytes. Our results revealed that peptide mixture inhibited melanosome biogenesis through the regulation of microphthalmia-associated transcription factor, a key factor of melanogenesis in melanocytes. And we observed that peptide mixture inhibited melanosome uptake through the reduction of protease-activated receptor 2, a phagocytosis-related receptor in keratinocytes. Furthermore, peptide mixture activated autophagy system resulting in degradation of transferred melanosomes in keratinocytes. The anti-pigmentation effect of multi-targeting peptide mixture was assessed in a human skin equivalent model (MelanoDerm). Melanin contents in epidermal layer were significantly decreased by topical treatment of peptide mixture, suggesting that it can be applied as a novel cosmetics material having a whitening function.
Melanin is produced by melanocytes located in the basal layer of the epidermis to protect skin cells against harmful external agents, including ultraviolet (UV) rays, fine dust, and various chemical compounds [1]. The factors secreted from keratinocytes mainly by UV irradiation, including alpha-melanocyte-stimulating hormone (α-MSH), adrenocorticotropic hormone, stem cell factor, endothelin 1, hepatocyte growth factor, basic fibroblast growth factor, and granulocyte-macrophage colony-stimulating factor, stimulate each receptors on the surface of melanocytes and melanin is produced in lysosome-related organelle called melanosome [2-7]. Among these factors, α-MSH is a small peptide hormone derived from pro-opiomelanocortin. Binding of α-MSH to melanocortin 1 receptor increases cyclic adenosine monophosphate (cAMP) level that activates protein kinase A (PKA) [8,9]. PKA then phosphorylates cAMP responsive element binding protein (CREB) and transcription of microphthalmia-associated transcription factor (MITF) is induced by phospho-CREB [10]. MITF acts as a transcription factor for tyrosinase, tyrosinase-related protein 1 (TYRP1), and dopachrome tautomerase (Dct or TYRP2), key enzymes in melanogenesis, and induces their expression [11]. The matured melanosome moves toward the dendrite tip, and melanosome motor protein complex consisting of Rab27a, Melanophilin, and Myosin Va plays a role in this process [12-14]. Melanophilin, which has binding domains for Rab27a, Myosin Va, and actin, simultaneously binds to Rab27a on the melanosome surface and to Myosin Va interacting with actin filament. The assembly of these motor proteins is required for melanosome transport to the dendrite tips along actin filament [15-19]. Subsequently, melanosomes transferred from dendrite tips of melanocytes to neighboring keratinocytes and incorporated melanosomes locate in the perinuclear area of keratinocytes [20,21]. Although proper melanin formation is important for normal cell protection, excessive melanin production induces hyperpigmentation, including melasma, freckles, and solar lentigo, which can lead to mental stress and poor quality of life [22,23]. Thus, there are demands for the development of satisfactory whitening agents without side effects such as skin irritation, allergy induction, and carcinogenesis and many studies targeting various whitening mechanisms have been conducted [24,25].
Peptides are short chain of amino acids, the smallest unit of protein, linked to each other by peptide bonds. Recently, extensive research and development regarding peptides as active ingredients have been performed in the cosmetics and pharmaceutical industries because of their high biocompatibility and protein mimicking ability [26,27].
We screened our peptide library and found four peptides having each effect on inhibition of melanin synthesis, migration and transfer as well as melanosome degradation promoting activity. Subsequently, this study aimed to investigate the whitening activity of a peptide mixture containing four peptides with the same molar ratio.
The 2-CTC resin used in the peptide synthesis was purchased from BeadTech (Seoul, Korea). Fmoc-amino acid, hydroxybenzotriazole (HOBt), and N,N,N´,N´-tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) were purchased from CS Bio Co. (San Francisco, CA, USA). The reagents used in the synthesis, including dimethylformamide (DMF), N,N-diisopropylethylamine (DIEA), piperidine, trifluoroacetic acid (TFA), thioanisole, phenol, ethane dithiol (EDT), triisopropylsilane (TIS), and diethyl ether, were purchased from Daejung Chemical & Metal Co., Ltd. (Seoul, Korea).
Dulbecco's Modified Eagle Medium (DMEM) powder for melanocytes and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (Waltham, MA, USA), and liquid DMEM media and penicillin/streptomycin were purchased from Welgene Inc. (Gyeongsan, Korea). Sodium bicarbonate, sodium hydroxide, arbutin, 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT), 3,4-dihydroxy-L-phenylalanine (L-DOPA), trypsin, and rapamycin were purchased from Sigma-Aldrich (St. Louis, MO, USA), and Solvable was purchased from PerkinElmer (Waltham, MA, USA). Antibodies against Rab27A, Melanophilin, MITF, Tyrosinase, and Actin were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), and antibodies against Beclin-1, LC3, p62, p-CREB and p-ERK1/2 were purchased from Cell Signaling Technology (Danvers, MA, USA). Primer synthesis was performed in Cosmo GENETECH (Seoul, Korea) and further used in the experiment (Table 1).
CTC resin with a capacity of 0.84 mmol/g was subjected to swelling reaction in a reactor containing DMF solvent. Then, 2 equivalents of Fmoc-C terminal amino acid and 2.5 equivalents of DIEA were added in DMF to the reactor and further subjected to reaction for 2 h. The termination of the reaction was confirmed with a Kaiser test kit (Sigma-Aldrich), and the resin was washed with DMF. The Fmoc was removed by adding 20% piperidine/DMF twice to the washed resin. The termination of the reaction was confirmed with the Kaiser test kit, and the resin was washed with DMF. Subsequently, the following process was repeated according to the sequence of amino acids in the C terminal to N terminal direction. After 2 equivalents of Fmoc-amino acid, 2 equivalents of HBTU, 2 equivalents of HOBt, and 2.5 equivalents of DIEA were added in DMF, the reaction was performed for 2 h. The termination of the reaction was confirmed with the Kaiser test kit, and the resin was washed with DMF. The Fmoc was removed from amino acid by adding 20% piperidine/DMF twice to the washed resin. The termination of the reaction was confirmed with the Kaiser test kit, and the resin was washed with DMF. In case of Pep-3, Ferulic acid conjugation was performed through the following procedure. After 2 equivalents of Ferulic acid, 2 equivalents of HBTU, 2 equivalents of HOBt, and 2.5 equivalents of DIEA were added in DMF, the reaction was performed for 2 h. The termination of the reaction was confirmed with the Kaiser test kit, and the resin was washed with DMF. After the final synthesis was completed, a cleavage solution (TFA:H₂O:thioanisole:phenol:EDT:TIS = 81.5:5:5:5:2.5:1) was added to separate the peptide from the resin. The peptide was then precipitated using diethyl ether and washed five times. Subsequently, drying was performed to recover the final peptide product.
The purity of the synthesized peptide was identified by HPLC (Thermo Fisher Scientific/U-3000) analysis, and it was detected at UV 214 nm under the flow rate of 1 ml/min under mobile phase 0.1% TFA in water/0.1% TFA in acetonitrile using a C18 (Agilent, Pursuit XRs, 250 × 4.6 mm, 5 μm, 100Å) column.
To identify the molecular weight, LC-MS/MS (AB SCIEX, 3200 Q-trap) analysis was performed, and it was detected by MS/MS at the flow rate of 0.25 ml/min under mobile phase 0.1% formic acid in water/0.1% formic acid in acetonitrile gradient using a C18 (Agilent, Pursuit XRs, 100 × 2.0 mm, 5 μm, 100Å) column. The MS/MS analysis conditions included ESI Positive mode, Source/Gas: CUR = 20, CAD = High, IS = 5500, TEM = 350, GS1 = 50, GS2 = 50/Compound DP = 50–80, EP = 10, CE = 10–50, and CES = 1–10 (Table 2).
The B16F10 melanoma cells used in this experiment were purchased from ATCC (Manassas, VA, USA). B16F10 cells were incubated in DMEM media containing 10% heat-inactivated FBS, 1% penicillin/streptomycin, and 1.5 g/L sodium bicarbonate at 37°C and under the condition of 5% CO2.
HaCaT keratinocytes were purchased from CLS (Eppelheim, Germany). HaCaT cells were incubated in DMEM media containing 10% heat-inactivated FBS and 1% penicillin/streptomycin at 37°C and under the condition of 5% CO2.
B16F10 cells were seeded in 48-well plate at a density of 8 × 103 cells/well and incubated for 16 hours. They were then treated with various concentrations of peptide mixture in 2% FBS containing media for 3 days. After incubation, 20 μl of 4 mg/ml MTT was treated to each well for 4 hours and the media was removed. Subsequently, cells were treated with 500 μl of DMSO to dissolve formazan. Absorbance was measured at 570 nm using a spectrophotometer (Molecular Devices, San Jose, CA, USA).
HaCaT cells were seeded in 96-well plate at a density of 5 × 103 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in serum-free media for 3 days. Subsequently, MTT assay was performed in the same manner as described above.
B16F10 cells were seeded in 6-well plate at a density of 5 × 104 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in 2% FBS containing media for 3 days. To induce melanin production, 100 ng/ml α-MSH was co-treated and 200 μM arbutin was used as a positive control. The cells were collected and lysed by treating with 200 μl of 1 M NaOH and absorbance for lysate at 490 nm was measured with a spectrophotometer.
B16F10 cells were seeded in 6-well plate at a density of 5 × 104 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in 2% FBS containing media for 3 days. To induce melanin production, 100 ng/ml α-MSH was co-treated and 200 μM arbutin was used as a positive control. Each well was treated with 200 μl of lysis buffer (Sigma-Aldrich) to collect the lysate. After quantitation of the protein using BCA kit (Thermo Fisher Scientific), 90 μg of protein from each group was incubated with 20 μl of 10 mM L-DOPA in 96-well plate for 30 min at 37°C. Absorbance for mixture at 475 nm was measured with a spectrophotometer.
B16F10 cells were seeded in 6-well plate at a density of 5 × 104 cells/well and incubated for 16 h. Cells were treated with 100 ng/ml α-MSH for 72 h to induce melanin synthesis and harvested by trypsin/EDTA treatment. Cells were then washed with 10 ml of 0.25 M sucrose (in 10 mM HEPES buffer) and resuspended in 10 ml of 0.25 M sucrose solution. Subsequently, after freezing with liquid nitrogen, thawing was performed at 37°C, which was repeated 10 times. After centrifugation at 1,000 g for 10 min, the supernatant was collected and further centrifuged for 45 min at 20,630 g and 4°C. Subsequently, the pellet was subjected to resuspension with DPBS and stored at –80°C before use.
HaCaT cells were seeded in 6-well plate at a density of 1 × 105 cells/well and incubated for16 h. They were then treated with the peptide mixture in serum-free media for 1 h and further treated with isolated melanosomes for 40 h. The cells were lysed by treating with 200 μl of 1 M NaOH and absorbance for lysate at 490 nm was measured with a spectrophotometer. Furthermore, melanosome staining was performed using a Fontana-Masson Stain Kit (ScyTek Laboratories, Logan, UT, USA) and images of cells were detected by light microscope.
HaCaT cells were seeded in 6-well plate at a density of 1 × 105 cells/well and incubated for 16 h. They were pretreated with isolated melanosomes in serum-free media for 48 h and additionally treated with the peptide mixture for 72 h. One micromole of rapamycin was used as positive control. The cells were lysed by treating with 200 μl of 1 M NaOH and absorbance for lysate at 490 nm was measured with a spectrophotometer. Furthermore, melanosome staining was performed using a Fontana-Masson Stain Kit and images of cells were detected by light microscope.
B16F10 cells were seeded in 6-well plate at a density of 5 × 104 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in 2% FBS containing media for 3 days. To induce melanin production, 100 ng/ml α-MSH was co-treated and 200 μM arbutin was used as a positive control. RNA was isolated from cells using RNA extraction kit (Qiagen, Germany), and cDNA synthesis was performed using RT-PCR premix (iNtRON Biotechnology, Seongnam, Korea). After preparation of reaction mixture with PCR premix (iNtRON Biotechnology) and primers for each gene, PCR was performed using PCR machine (Eppendorf, Germany). Subsequently, mRNA expression patterns were determined by agarose gel electrophoresis.
HaCaT cells were seeded in 6-well plate at a density of 3 × 105 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in the serum-free media for 1 h and additionally treated with 4U trypsin for 16 h. The cells were collected and RT-PCR was performed in the same manner as described above.
B16F10 cells were seeded in 6-well plate at a density of 5 × 104 cells/well and incubated for 1 day. They were then treated with various concentrations of peptide mixture in 2% FBS containing media for 3 days. To induce melanin production, 100 ng/ml α-MSH was co-treated and 200 μM arbutin was used as a positive control. The cells were lysed with lysis buffer and the protein was quantified using BCA kit. Twenty micrograms of protein were separated on 8% polyacrylamide gel and electrophoretically transferred to polyvinylidene fluoride membranes. Membranes were blocked in 5% w/v skim milk in PBS with 0.5% Tween 20 (PBS-T). Incubation with primary antibodies diluted in blocking solution was performed overnight at 4°C and was followed by washing with PBS-T. The appropriate secondary antibodies were diluted in blocking solutions and incubated with the membranes for 1 h at room temperature followed by washing with PBS-T. The membranes were visualized via Western detection reagent (Elpis Biotech, Daejeon, Korea) with Gel Doc (Bio-Rad, Hercules, CA, USA).
For analysis of p-CREB and p-ERK, B16F10 cells were seeded in 6-well plate at a density of 3 × 105 cells/well and incubated for 1 day. They were then treated with various concentrations of peptide mixture in 2% FBS containing media. The incubation times were 30 min for analysis of p-CREB and 10 min for analysis of p-ERK. To induce melanin production, 100 ng/ml α-MSH was co-treated and 200 μM arbutin was used as a positive control. Western blotting was performed using isolated proteins in the same manner as described above.
HaCaT cells were seeded in 6-well plate at a density of 3 × 105 cells/well and incubated for 16 h. They were then treated with various concentrations of peptide mixture in serum-free media for 3 days. Two hundred nanograms of rapamycin was treated for 3 hours as a positive control. Western blotting was performed using isolated proteins in the same manner as described above.
To test the whitening effect of peptide mixture using skin equivalent, liposome sample was prepared as follows. The peptide mixture was dissolved in distilled water at concentration of 2,000 μg/ml and adjusted to pH 7. After filtration using 0.22-μm pore size filter, 1% hydrogenated phosphatidylcholine buffer was mixed at a ratio of 10%. The nano-sized liposome sample was prepared by passing through the 75-μm cells five times at 1,000 bar using the Microfluidizer.
MelanoDerm MEL-300-B (MatTek Corporation, Ashland, MA, USA) was topically treated with 25 μl or 50 μl of liposome containing 2,000 μg/ml peptide mixture three times a week for 2 weeks. Same volume of vehicle was applied to control tissue. The treated skin equivalents were washed with DPBS and light microscopy was performed to check morphology of melanocytes. Subsequently, to analysis the production of melanin, skin equivalents were frozen at –80°C for 30 min and thawed at room temperature. Tissue samples were incubated with 1% sodium bicarbonate for 30 min and dried. After treatment with 300 μl of Solvable, the samples were left overnight at 95°C and then cooled at room temperature. Absorbance of the tissue extract at 490 nm was measured with a spectrophotometer.
To observe the melanin distribution within the tissues through histology, all samples were fixed in 4% phosphate-buffered formalin for 24 h. Fixed samples were paraffin-embedded and cut into a 5-μm section using microtome (Leica Biosystems, Wetzlar, Germany). Paraffin sections were stained using Fontana-Masson Stain Kit according to the manufacturer's instructions and light microscopy were performed.
The experiments conducted in this study were repeated three times or more. The statistical significance of the data was tested through Student's t-test. The resulting values were expressed as mean ± standard deviation and were considered statistically significant when the p value was less than 0.05.
To determine the effect of the peptide mixture on cell viability, B16F10 melanoma cells and HaCaT keratinocytes were treated with the peptide mixture at concentrations of 10, 50, 100, and 200 μM, and MTT assay was performed. The results showed that peptide mixture was not cytotoxic in the overall treatment concentrations, and subsequent experiments were performed in the above concentration range (Fig. 1A).
To identify the inhibitory effect of the peptide mixture on melanin synthesis, B16F10 cells were treated at concentrations of 10, 50, 100, and 200 μM to compare melanin production rates. The results showed dose-dependent inhibition of melanin production by the peptide mixture. Inhibition rates were 3% at the lowest concentration of 10 μM and 26% at the highest concentration of 200 μM (Fig. 1B).
Tyrosinase is one of the key enzymes in melanin synthesis. The tyrosinase activity assay was performed to verify that the melanogenesis inhibitory effect of peptide mixture was caused by the regulation of tyrosinase. The results showed significant inhibition of tyrosinase activity by the peptide mixture in B16F10 cells. Inhibition rates were 8% at the lowest concentration of 10 μM and 33% at the highest concentration of 200 μM (Fig. 1C).
To investigate the mechanism of depigmenting action shown by peptide mixture, expression levels of melanogenesis-related factors and melanosome transportation-related factors were analyzed.
As shown in Fig. 2A, transcription levels of MITF and of its downstream genes, TYR, TYPR1 and TYRP2, were decreased by peptide mixture in B16F10 cells. Furthermore, α-MSH-induced protein levels of MITF and Tyrosinase were significantly decreased by peptide mixture in B16F10 cells (Fig. 2B).
These results suggest that the melanogenesis inhibition effect of peptide mixture occurred by inhibiting the expression of melanogenesis-related factors.
It has been published that MITF also acts as a transcription factor that activates motor proteins which have role in melanosome transportation [28,29]. To identify whether the peptide mixture, which has inhibitory effect on MITF expression, decreased the expression of motor proteins, we analyzed mRNA and protein levels. As shown in Fig. 2C, transcription levels of RAB27A, MLPH, and MYO5A genes were significantly decreased by peptide mixture in B16F10 cells. Furthermore, α-MSH-induced protein levels of Rab27a and Melanophilin were decreased by peptide mixture in B16F10 cells (Fig. 2D).
These results suggest that inhibition of MITF expression by peptide mixture can be result in inhibition of melanosome transportation.
To investigate the action mechanism of peptide mixture on MITF expression, we analyzed phosphorylation level of transcriptional and post-translational regulatory factors of MITF.
As shown in Fig. 3A, phosphorylated CREB, a transcription factor activating MITF expression, was dose-dependently decreased by peptide mixture. Previously, it has been identified that activity of MITF depends on its post-translational modifications and proteasome-mediated MITF protein degradation can be induced by phosphorylated ERK1/2 [30]. Peptide mixture significantly increased p-ERK1/2 level under α-MSH-stimulated condition in B16F10 cells (Fig. 3B).
These results suggest that the expression and activity of MITF might be decreased through the regulation of CREB and ERK1/2 phosphorylations by peptide mixture.
Melanosomes are known to be transferred from dendrite tips of melanocytes to keratinocytes through the phagocytosis of keratinocytes. To identify whether the peptide mixture could inhibit the melanosome transfer to keratinocytes, HaCaT keratinocytes were treated with the peptide mixture at concentrations of 10, 50, 100, and 200 μM and further treated with melanosomes isolated from B16F10 cells. The amount of melanosomes transferred to the cells was dose-dependently decreased by peptide mixture (Fig. 4A). Image analysis through Fontana-Masson staining showed that the distribution of brown-stained melanosomes significantly decreased in the peptide mixture-treated group in a dose-dependent manner (Fig. 4B).
Furthermore, the expression of F2RL1 (PAR-2) gene, a receptor that regulates the phagocytosis of keratinocytes, was significantly decreased by peptide mixture under trypsin-stimulated condition (Fig. 4C).
These results suggest that the peptide mixture can reduce phagocytosis through the inhibition of PAR-2 expression in keratinocytes by the peptide mixture.
Melanosomes transferred into keratinocytes are known to be gradually eliminated by the cornification of keratinocytes or degraded by intracellular autophagy system. To identify whether the peptide mixture could promote the melanosome degradation, we treated peptide mixture to the melanosome containing HaCaT keratinocytes. When the amounts of melanosomes remaining was monitored, dose-dependent decrease was observed from the cell lysate treated with peptide mixture (Fig. 5A). Image analysis through Fontana-Masson staining showed that the distribution of brown-stained melanosomes significantly decreased in the peptide mixture-treated group in a dose-dependent manner (Fig. 5B).
Furthermore, when the autophagic activity related-protein levels in keratinocytes were monitored, the protein levels of Beclin-1 and LC3-II in relation to autophagosome formation were increased by peptide mixture treatment, while the protein level of p62 degraded by autophagolysosomes was decreased by peptide mixture treatment (Fig. 5C).
These results suggest that the peptide mixture induced the autophagic activity to promote the degradation of the melanosomes in keratinocytes.
An experiment using MelanoDerm was conducted to identify whether the peptide mixture could result in anti-pigmentation of the skin. After topical treatment of liposome containing 2,000 ppm of the peptide mixture was conducted at doses of 50 and 100 μg for 2 weeks, light microscopy was performed. The formation of dark melanosomes was induced entirely in the melanocyte bodies and dendrite tips of the control tissue, while the overall cell bodies were bright with a smaller number of dendrites because melanin production was inhibited in the tissue treated with the peptide mixture (Fig. 6A). Also, the melanin contents were dose-dependently decreased by peptide mixture, supporting the visual observation (Fig. 6B).
Moreover, histology was performed to observe the melanin distribution throughout the epidermis. According to the result, the control tissue showed a marked increase of melanogenesis in melanocytes of the basal layer, and brown-stained melanin was observed in keratinocytes due to the accompanied transfer to nearby keratinocytes. A considerable level of melanin was also observed in the upper cornified layers. On the other hand, the melanin contents of melanocytes were low in the tissues treated with the peptide mixture, and a significant decrease in melanin contents were observed throughout keratinocytes and cornified layers in comparison to the control group (Fig. 6C).
These results confirmed a significant anti-pigmentation effect of the peptide mixture on the skin equivalents.
The development of whitening materials has been targeted mainly to inhibit melanogenesis in melanocytes through the regulation of melanogenesis-related factors [31-35]. Recently, other targets of depigmentation, including melanosome migration, transfer and degradation, have been studied and several agents have been identified [36-42]. Because pigmentation is an action involving multiple mechanism, it might be efficient to use the combination of therapeutic agents which have regulatory effects for each target.
This study investigated the whitening activity of a peptide mixture containing four different peptides with the same molar ratio. The suggested action mechanism of peptide mixture against melanosome biogenesis, transfer and degradation is illustrated in Fig. 7. In melanocytes, the peptide mixture inhibits CREB phosphorylation results in decreased MITF expression and induces phosphorylation of ERK1/2 results in MITF phosphorylation and proteasomal degradation. As a result, the expression of melanosome biogenesis and transport-related proteins, including Tyrosinase, TYRP1, TYRP2, Rab27A, Melanophilin and MYO5A, are decreased. In keratinocytes, the peptide mixture decreases expression of PAR-2 that mediates keratinocyte phagocytic capability correlated with actin filament reorganization. Furthermore, the peptide mixture activates autophagic flux, and transferred melanosomes are degraded in autophagolysosome.
Peptides have been extensively examined as active ingredients for cosmetics because of their high biocompatibility and protein-mimicking activity [43]. But they also have some disadvantages that they can be easily degraded by proteases present in the skin and cannot easily penetrate the epidermis composed of lipids due to their hydrophilicity [44]. The studies for increasing peptide stability have suggested the methods of substituting amino acids at predicted cleavage site, acetylation of the N terminal, and amidation of the C terminal [45]. Furthermore, there are many studies to improve the transdermal penetration of peptides: methods involving the use of penetration inducers such as alcohols, azones, hexanoates, and unsaturated fatty acids; methods involving the use of specific peptide sequences that facilitate permeation; methods combining lipophilic derivatives; and methods encapsulating peptides such as liposomes, transfersomes, niosomes, and ethosomes [46-48]. In this regard, we prepared liposome encapsulating the peptide mixture to test its whitening effect in skin equivalent model by topical treatment, and significant reduction of melanin contents throughout the epidermal layers was shown in the test liposome treatment group (Fig. 6C).
In conclusion, the peptide mixture of this study exhibited whitening effect through various actions, including inhibition of melanin synthesis and migration as well as inhibition of melanosome uptake to keratinocytes and promotion of melanosome degradation. The peptide mixture of this study could be used as a new whitening material and that the liposome that facilitates the stability and skin penetration of peptides could be used to develop effective whitening products.
ACKNOWLEDGEMENTS
This study was conducted under the support of the World Class 300 R&D Project by the Ministry of Trade, Industry, and Energy (Project number: S2641452).
Notes
REFERENCES
1. Hirobe T. 2014; Keratinocytes regulate the function of melanocytes. Dermatol Sin. 32:200–204. DOI: 10.1016/j.dsi.2014.05.002.

2. Hachiya A, Kobayashi A, Ohuchi A, Takema Y, Imokawa G. 2001; The paracrine role of stem cell factor/c-kit signaling in the activation of human melanocytes in ultraviolet-B-induced pigmentation. J Invest Dermatol. 116:578–586. DOI: 10.1046/j.1523-1747.2001.01290.x. PMID: 11286626.


3. Hirobe T, Hasegawa K, Furuya R, Fujiwara R, Sato K. 2013; Effects of fibroblast-derived factors on the proliferation and differentiation of human melanocytes in culture. J Dermatol Sci. 71:45–57. DOI: 10.1016/j.jdermsci.2013.03.012. PMID: 23726358.


4. Schauer E, Trautinger F, Köck A, Schwarz A, Bhardwaj R, Simon M, Ansel JC, Schwarz T, Luger TA. 1994; Proopiomelanocortin-derived peptides are synthesized and released by human keratinocytes. J Clin Invest. 93:2258–2262. DOI: 10.1172/JCI117224. PMID: 8182158. PMCID: PMC294380.



5. Chakraborty AK, Funasaka Y, Slominski A, Ermak G, Hwang J, Pawelek JM, Ichihashi M. 1996; Production and release of proopiomelanocortin (POMC) derived peptides by human melanocytes and keratinocytes in culture: regulation by ultraviolet B. Biochim Biophys Acta. 1313:130–138. DOI: 10.1016/0167-4889(96)00063-8. PMID: 8781560.

6. Wakamatsu K, Graham A, Cook D, Thody AJ. 1997; Characterisation of ACTH peptides in human skin and their activation of the melanocortin-1 receptor. Pigment Cell Res. 10:288–297. DOI: 10.1111/j.1600-0749.1997.tb00688.x. PMID: 9359624.


7. Imokawa G. 2004; Autocrine and paracrine regulation of melanocytes in human skin and in pigmentary disorders. Pigment Cell Res. 17:96–110. DOI: 10.1111/j.1600-0749.2003.00126.x. PMID: 15016298.


8. Chakraborty A, Slominski A, Ermak G, Hwang J, Pawelek J. 1995; Ultraviolet B and melanocyte-stimulating hormone (MSH) stimulate mRNA production for alpha MSH receptors and proopiomelanocortin-derived peptides in mouse melanoma cells and transformed keratinocytes. J Invest Dermatol. 105:655–659. DOI: 10.1111/1523-1747.ep12324134. PMID: 7594638.

9. Khaled M, Larribere L, Bille K, Aberdam E, Ortonne JP, Ballotti R, Bertolotto C. 2002; Glycogen synthase kinase 3beta is activated by cAMP and plays an active role in the regulation of melanogenesis. J Biol Chem. 277:33690–33697. DOI: 10.1074/jbc.M202939200. PMID: 12093801.
10. Kim YM, Cho SE, Seo YK. 2016; The activation of melanogenesis by p-CREB and MITF signaling with extremely low-frequency electromagnetic fields on B16F10 melanoma. Life Sci. 162:25–32. DOI: 10.1016/j.lfs.2016.08.015. PMID: 27543340.


11. Kameyama K, Sakai C, Kuge S, Nishiyama S, Tomita Y, Ito S, Wakamatsu K, Hearing VJ. 1995; The expression of tyrosinase, tyrosinase-related proteins 1 and 2 (TRP1 and TRP2), the silver protein, and a melanogenic inhibitor in human melanoma cells of differing melanogenic activities. Pigment Cell Res. 8:97–104. DOI: 10.1111/j.1600-0749.1995.tb00648.x. PMID: 7659683.


12. Van Gele M, Geusens B, Schmitt AM, Aguilar L, Lambert J. 2008; Knockdown of myosin Va isoforms by RNAi as a tool to block melanosome transport in primary human melanocytes. J Invest Dermatol. 128:2474–2484. DOI: 10.1038/jid.2008.100. PMID: 18401430.


13. Ohbayashi N, Fukuda M. 2012; Role of Rab family GTPases and their effectors in melanosomal logistics. J Biochem. 151:343–351. DOI: 10.1093/jb/mvs009. PMID: 22323658.

14. Park JI, Lee HY, Lee JE, Myung CH, Hwang JS. 2016; Inhibitory effect of 2-methyl-naphtho[1,2,3-de]quinolin-8-one on melanosome transport and skin pigmentation. Sci Rep. 6:29189. DOI: 10.1038/srep29189. PMID: 27381646. PMCID: PMC4933902.

15. Oberhofer A, Spieler P, Rosenfeld Y, Stepp WL, Cleetus A, Hume AN, Mueller-Planitz F, Ökten Z. 2017; Myosin Va's adaptor protein melanophilin enforces track selection on the microtubule and actin networks in vitro. Proc Natl Acad Sci U S A. 114:E4714–E4723. DOI: 10.1073/pnas.1619473114. PMID: 28559319. PMCID: PMC5474823.

16. Provance DW, James TL, Mercer JA. 2002; Melanophilin, the product of the leaden locus, is required for targeting of myosin-Va to melanosomes. Traffic. 3:124–132. DOI: 10.1034/j.1600-0854.2002.030205.x. PMID: 11929602. PMCID: PMC1351229.



17. Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. 2002; A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J Biol Chem. 277:25423–25430. DOI: 10.1074/jbc.M202574200. PMID: 11980908.
18. Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA 3rd. 2002; Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 4:271–278. DOI: 10.1038/ncb760. PMID: 11887186.


19. Kuroda TS, Ariga H, Fukuda M. 2003; The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol Cell Biol. 23:5245–5255. DOI: 10.1128/MCB.23.15.5245-5255.2003. PMID: 12861011. PMCID: PMC165717.

20. Cardinali G, Ceccarelli S, Kovacs D, Aspite N, Lotti LV, Torrisi MR, Picardo M. 2005; Keratinocyte growth factor promotes melanosome transfer to keratinocytes. J Invest Dermatol. 125:1190–1199. DOI: 10.1111/j.0022-202X.2005.23929.x. PMID: 16354189.


21. Epstein JH. 1983; Photocarcinogenesis, skin cancer, and aging. J Am Acad Dermatol. 9:487–502. DOI: 10.1016/S0190-9622(83)70160-X. PMID: 6355213.


22. Speeckaert R, Van Gele M, Speeckaert MM, Lambert J, van Geel N. 2014; The biology of hyperpigmentation syndromes. Pigment Cell Melanoma Res. 27:512–524. DOI: 10.1111/pcmr.12235. PMID: 24612852.


23. Maymone MBC, Neamah HH, Secemsky EA, Vashi NA. 2018; Correlating the Dermatology Life Quality Index and Skin Discoloration Impact Evaluation Questionnaire tools in disorders of hyperpigmentation. J Dermatol. 45:361–362. DOI: 10.1111/1346-8138.14172. PMID: 29498101.


24. Pillaiyar T, Manickam M, Namasivayam V. 2017; Skin whitening agents: medicinal chemistry perspective of tyrosinase inhibitors. J Enzyme Inhib Med Chem. 32:403–425. DOI: 10.1080/14756366.2016.1256882. PMID: 28097901. PMCID: PMC6010116.



25. Juhasz MLW, Levin MK. 2018; The role of systemic treatments for skin lightening. J Cosmet Dermatol. 17:1144–1157. DOI: 10.1111/jocd.12747. PMID: 30133125.


26. Zhang L, Falla TJ. 2009; Cosmeceuticals and peptides. Clin Dermatol. 27:485–494. DOI: 10.1016/j.clindermatol.2009.05.013. PMID: 19695481.


27. Reddy B, Jow T, Hantash BM. 2012; Bioactive oligopeptides in dermatology: part I. Exp Dermatol. 21:563–568. DOI: 10.1111/j.1600-0625.2012.01528.x. PMID: 22672743.


28. Chiaverini C, Beuret L, Flori E, Busca R, Abbe P, Bille K, Bahadoran P, Ortonne JP, Bertolotto C, Ballotti R. 2008; Microphthalmia-associated transcription factor regulates RAB27A gene expression and controls melanosome transport. J Biol Chem. 283:12635–12642. DOI: 10.1074/jbc.M800130200. PMID: 18281284.

29. Alves CP, Yokoyama S, Goedert L, Pontes CLS, Sousa JF, Fisher DE, Espreafico EM. 2017; MYO5A gene is a target of MITF in melanocytes. J Invest Dermatol. 137:985–989. DOI: 10.1016/j.jid.2016.11.026. PMID: 27939378.

30. Hartman ML, Czyz M. 2015; MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 72:1249–1260. DOI: 10.1007/s00018-014-1791-0. PMID: 25433395. PMCID: PMC4363485.

31. Lajis AFB, Ariff AB. 2019; Discovery of new depigmenting compounds and their efficacy to treat hyperpigmentation: evidence from in vitro study. J Cosmet Dermatol. 18:703–727. DOI: 10.1111/jocd.12900. PMID: 30866156.


32. Garcia-Jimenez A, Teruel-Puche JA, Berna J, Rodriguez-Lopez JN, Tudela J, Garcia-Canovas F. 2017; Action of tyrosinase on alpha and beta-arbutin: a kinetic study. PLoS One. 12:e0177330. DOI: 10.1371/journal.pone.0177330. PMID: 28493937. PMCID: PMC5426667.

33. Yu JS, Kim AK. 2010; Effect of combination of taurine and azelaic acid on antimelanogenesis in murine melanoma cells. J Biomed Sci. 17(Suppl 1):S45. DOI: 10.1186/1423-0127-17-S1-S45. PMID: 20804622. PMCID: PMC2994384.

34. Lee CS, Jang WH, Park M, Jung K, Baek HS, Joo YH, Park YH, Lim KM. 2013; A novel adamantyl benzylbenzamide derivative, AP736, suppresses melanogenesis through the inhibition of cAMP-PKA-CREB-activated microphthalmia-associated transcription factor and tyrosinase expression. Exp Dermatol. 22:762–764. DOI: 10.1111/exd.12248. PMID: 24107097.


35. Huang HC, Chang SJ, Wu CY, Ke HJ, Chang TM. 2014; [6]-Shogaol inhibits α-MSH-induced melanogenesis through the acceleration of ERK and PI3K/Akt-mediated MITF degradation. Biomed Res Int. 2014:842569. DOI: 10.1155/2014/842569. PMID: 25045707. PMCID: PMC4090493.
36. Kim B, Lee JY, Lee HY, Nam KY, Park J, Lee SM, Kim JE, Lee JD, Hwang JS. 2013; Hesperidin suppresses melanosome transport by blocking the interaction of Rab27A-melanophilin. Biomol Ther (Seoul). 21:343–348. DOI: 10.4062/biomolther.2013.032. PMID: 24244821. PMCID: PMC3825197.



37. Chang H, Choi H, Joo KM, Kim D, Lee TR. 2012; Manassantin B inhibits melanosome transport in melanocytes by disrupting the melanophilin-myosin Va interaction. Pigment Cell Melanoma Res. 25:765–772. DOI: 10.1111/pcmr.12002. PMID: 22863119.


38. Makino-Okamura C, Niki Y, Takeuchi S, Nishigori C, Declercq L, Yaroch DB, Saito N. 2014; Heparin inhibits melanosome uptake and inflammatory response coupled with phagocytosis through blocking PI3k/Akt and MEK/ERK signaling pathways in human epidermal keratinocytes. Pigment Cell Melanoma Res. 27:1063–1074. DOI: 10.1111/pcmr.12287. PMID: 24961476.


39. Seiberg M, Paine C, Sharlow E, Andrade-Gordon P, Costanzo M, Eisinger M, Shapiro SS. 2000; The protease-activated receptor 2 regulates pigmentation via keratinocyte-melanocyte interactions. Exp Cell Res. 254:25–32. DOI: 10.1006/excr.1999.4692. PMID: 10623462.


40. Murase D, Hachiya A, Takano K, Hicks R, Visscher MO, Kitahara T, Hase T, Takema Y, Yoshimori T. 2013; Autophagy has a significant role in determining skin color by regulating melanosome degradation in keratinocytes. J Invest Dermatol. 133:2416–2424. DOI: 10.1038/jid.2013.165. PMID: 23558403.


41. Kim ES, Shin JH, Seok SH, Kim JB, Chang H, Park SJ, Jo YK, Choi ES, Park JS, Yeom MH, Lim CS, Cho DH. 2013; Autophagy mediates anti-melanogenic activity of 3'-ODI in B16F1 melanoma cells. Biochem Biophys Res Commun. 442:165–170. DOI: 10.1016/j.bbrc.2013.11.048. PMID: 24269817.

42. Li L, Chen X, Gu H. 2016; The signaling involved in autophagy machinery in keratinocytes and therapeutic approaches for skin diseases. Oncotarget. 7:50682–50697. DOI: 10.18632/oncotarget.9330. PMID: 27191982. PMCID: PMC5226613.



43. Schagen SK. 2017; Topical peptide treatments with effective anti-aging results. Cosmetics. 4:16. DOI: 10.3390/cosmetics4020016.

44. Reddy BY, Jow T, Hantash BM. 2012; Bioactive oligopeptides in dermatology: part II. Exp Dermatol. 21:569–575. DOI: 10.1111/j.1600-0625.2012.01527.x. PMID: 22672721.


45. Strömstedt AA, Pasupuleti M, Schmidtchen A, Malmsten M. 2009; Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob Agents Chemother. 53:593–602. DOI: 10.1128/AAC.00477-08. PMID: 19029324. PMCID: PMC2630634.

46. Pai VV, Bhandari P, Shukla P. 2017; Topical peptides as cosmeceuticals. Indian J Dermatol Venereol Leprol. 83:9–18. DOI: 10.4103/0378-6323.186500. PMID: 27451932.


47. Marepally S, Boakye CH, Shah PP, Etukala JR, Vemuri A, Singh M. 2013; Design, synthesis of novel lipids as chemical permeation enhancers and development of nanoparticle system for transdermal drug delivery. PLoS One. 8:e82581. DOI: 10.1371/journal.pone.0082581. PMID: 24349315. PMCID: PMC3861410.

48. Kalluri H, Banga AK. 2011; Transdermal delivery of proteins. AAPS PharmSciTech. 12:431–441. DOI: 10.1208/s12249-011-9601-6. PMID: 21369712. PMCID: PMC3066337.



Fig. 1
Effects of the peptide mixture on cell viability and melanogenesis.
(A) B16F10 melanoma cells and HaCaT keratinocytes were treated with indicated concentration of peptide mixture for 72 h. Cell viability was determined by MTT assay. (B) B16F10 cells were exposed to peptide mixture in the presence of 100 ng/ml α-MSH for 72 h. Melanin content and (C) cellular tyrosinase activity were measured. Two hundreds micromole of arbutin was used as a positive control. Each determination was made in triplicate, and data are shown as means ± standard deviation. MTT, 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide; α-MSH, alpha-melanocyte-stimulating hormone. *p < 0.05, **p < 0.01 compared to the α-MSH-treated control.
Fig. 2
Effects of the peptide mixture on the expression of melanogenesis-related factors and melanosome transport-related factors.
B16F10 cells were exposed to peptide mixture in the presence of 100 ng/ml α-MSH for 72 h. Two hundreds micromole of arbutin was used as a positive control. (A) Expression levels of melanogenesis-related genes including MITF, TYR, TYRP1, and TYRP2 were analyzed by RT-PCR. (B) The protein levels of MITF and tyrosinase were detected using Western blotting. (C) Expression levels of melanosome transport-related genes including RAB27A, MLPH, and MYO5A were analyzed by RT-PCR. (D) The protein levels of Rab27a and melanophilin were detected using Western blotting. α-MSH, alpha-melanocyte-stimulating hormone; MITF, microphthalmia-associated transcription factor; TYRP, tyrosinase-related protein.
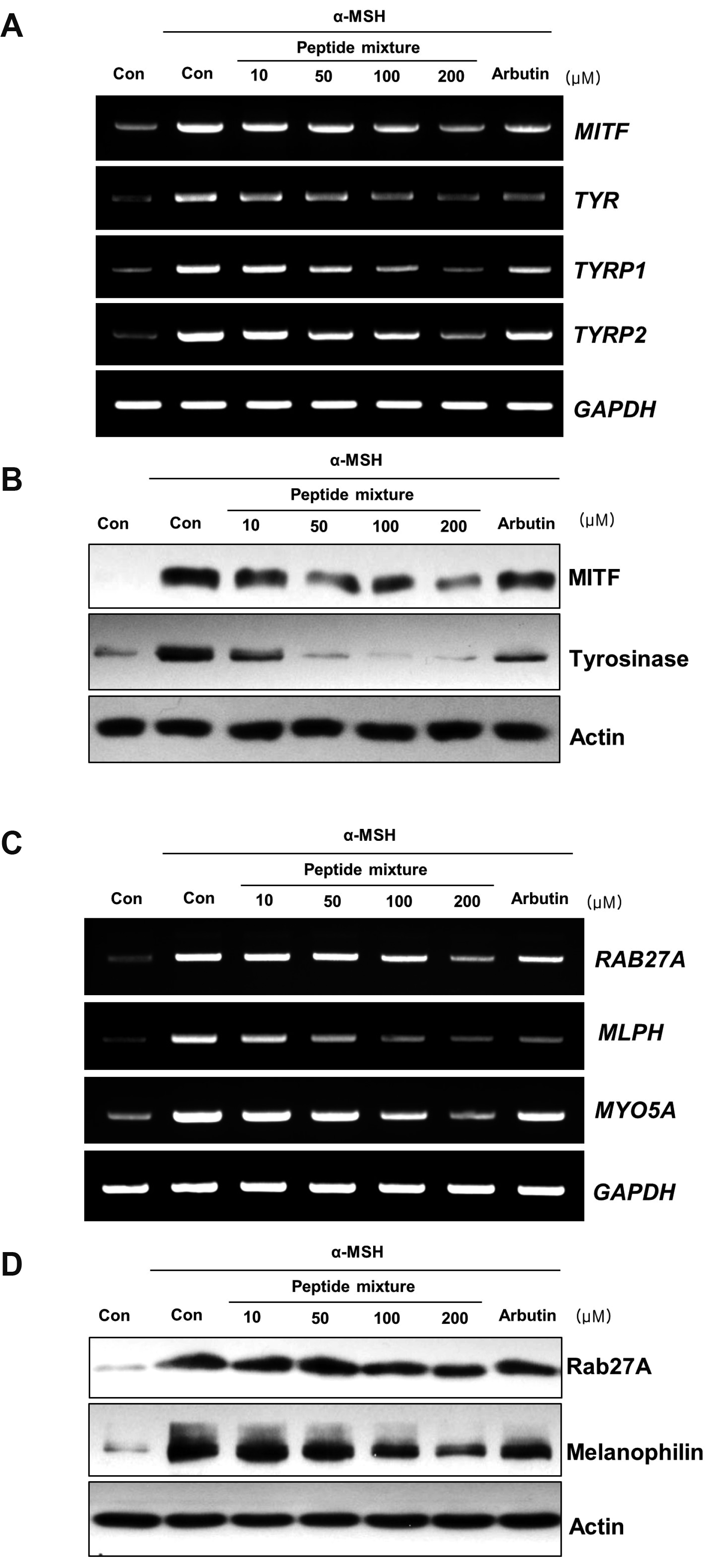
Fig. 3
Effects of the peptide mixture on the phosphorylation level of CREB and ERK.
(A) B16F10 cells were exposed to peptide mixture in the presence of 100 ng/ml α-MSH for 30 min. Two hundreds micromole of arbutin was used as a positive control. Phosphorylation level of CREB, a transcriptional factor of MITF, was analyzed by Western blotting. (B) B16F10 cells were exposed to peptide mixture in the presence of 100 ng/ml α-MSH for 10 min. Two hundreds micromole of arbutin was used as a positive control. Phosphorylation level of ERK, a kinase regulates protein stability of MITF, was analyzed by Western blotting. CREB, cAMP responsive element binding protein; α-MSH, alpha-melanocyte-stimulating hormone; MITF, microphthalmia-associated transcription factor.
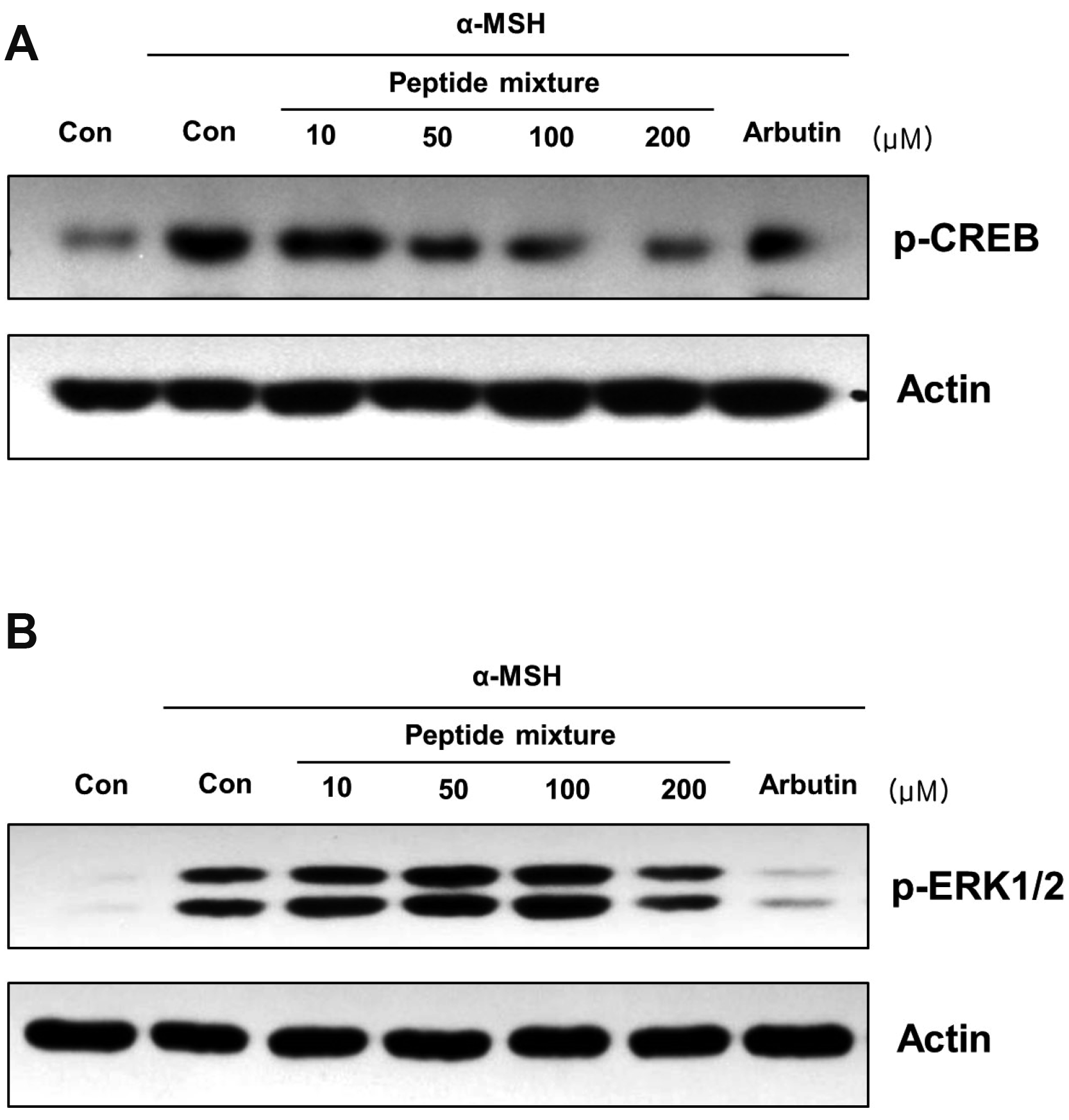
Fig. 4
Effects of the peptide mixture on the melanosome uptake of keratinocytes.
HaCaT keratinocytes were exposed to peptide mixture for 1 h and additionally incubated with isolated melanosomes for 40 h. (A) After treatment, melanin content was analyzed. Each determination was made in triplicate, and data are shown as means ± standard deviation. *p < 0.05, **p < 0.01 compared to the melanosome-treated control. (B) Fontana-Masson staining was performed to observe the intracellular distribution of melanosomes. (C) HaCaT keratinocytes were pretreated with the peptide mixture for 1 h, cells were additionally incubated with 4U trypsin for 16 h. The expression level of F2RL1 gene was analyzed by RT-PCR.
Fig. 5
Effect of the peptide mixture on melanosome degradation in keratinocytes.
HaCaT keratinocytes were exposed to isolated melanosomes for 48 h and additionally incubated with peptide mixture for 72 h. One micromole of rapamycin was used as a positive control. (A) After treatment, melanin content was analyzed. Each determination was made in triplicate, and data are shown as means ± standard deviation. *p < 0.05 compared to the melanosome-treated control. (B) Fontana-Masson staining was performed to observe the intracellular distribution of melanosomes. (C) HaCaT keratinocytes were treated with peptide mixture for 3 h and protein levels of Beclin-1, LC3, and p62 were analyzed by western blotting. 200 nM rapamycin was used as a positive control.

Fig. 6
The whitening effect of the peptide mixture on the skin equivalent.
MelanoDerm (MEL-300-B) was topically treated with liposome containing 2,000 μg/ml of peptide mixture three times a week for 2 weeks. (A) Light microscopy was conducted to observe the cell morphology (B) and melanin content was analyzed using extract of equivalent. (C) Paraffin sections were stained with Fontana-Masson to observe the melanosome distribution of the epidermal layers (scale bar = 100 μm). Each determination was made in triplicate, and data shown are means ± standard deviation. *p < 0.05 compared to the untreated control.

Fig. 7
Schematic diagram showing the mechanism of action of peptide mixture.
CREB, cAMP-response-element-binding protein; MITF, microphthalmia-associated transcription factor; TYRP1, tyrosinase-related protein 1; TYRP2, tyrosinase-related protein 2; ERK, extracellular signal regulated kinase; PAR-2, protease activated receptor 2; PE, phosphatidylethanolamine. Arrows indicate positive regulation while T-bars denote inhibitory effects.
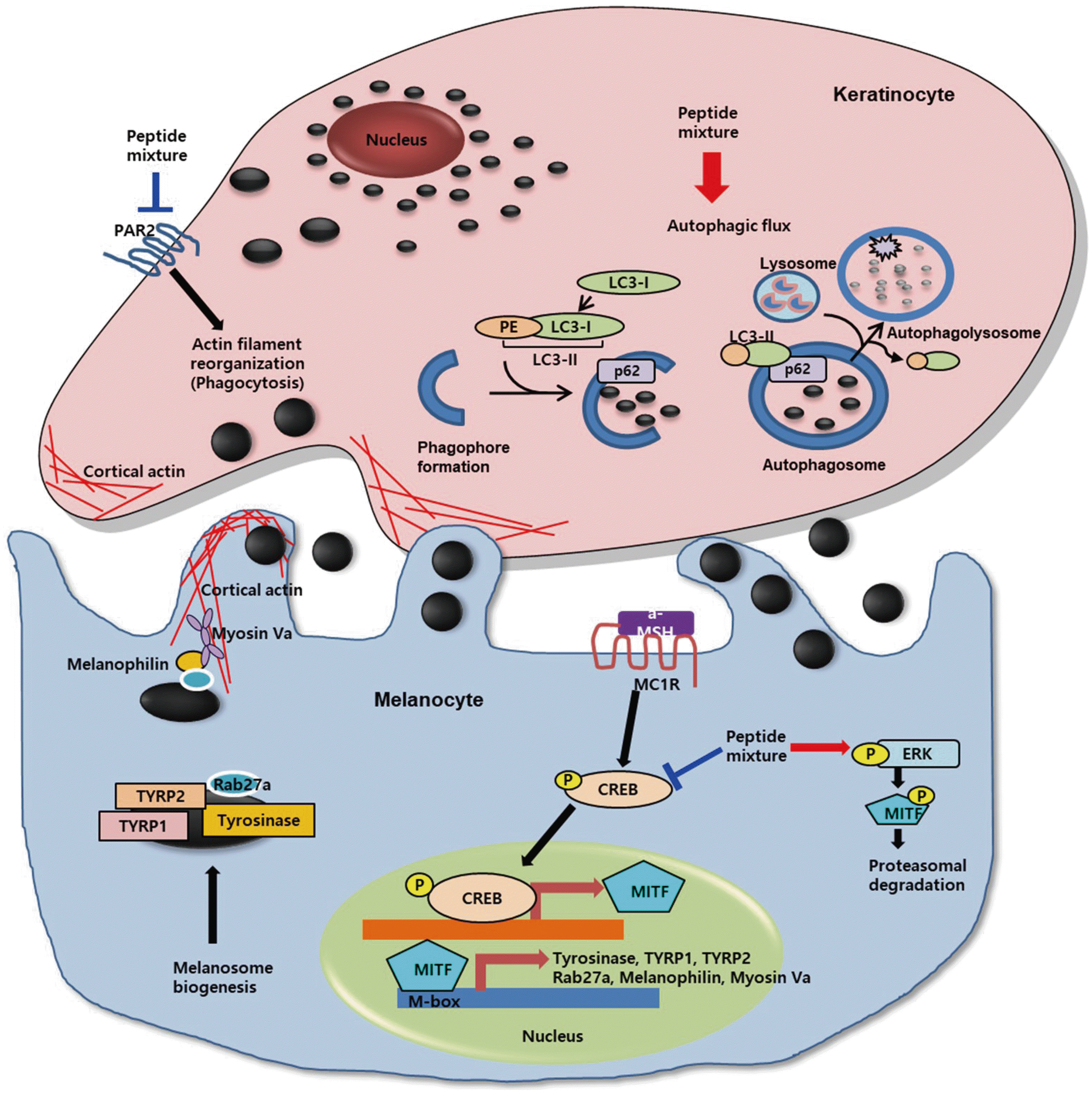
Table 1
Primer sequences




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download