INTRODUCTION
METHODS
Reagents
Animal model
Administration
Determination of lung function A catheter is inserted into the trachea of the body
Histopathology and immunohistochemistry
Western blotting
Cell line and culture
Transfection
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Statistical analysis
RESULTS
Association between Nrf2 and pathogenesis of CS-induced COPD
 | Fig. 1Pathological evaluation of lung tissue.(A) Changes in lung histopathology observed following H&E staining. (B) Average score of airway inflammation. #p < 0.05 vs. control group, △p < 0.05 vs. model group, *p < 0.05 vs. WT model group (n = 3). (C) Destructive index of alveoli, #p < 0.05 vs. control group, *p < 0.05 vs. model group, △p < 0.05 vs. WT model group (n = 3). WT, wild type; AITC, allyl isothiocyanate; NAC, N-acetylcysteine; Nrf2, nuclear factor erythroid 2-related factor 2.
|
Table 1
| Group | FEV0.3/FVC% | FEV0.1 | PEF | FEF25–75 | Cdyn |
|---|---|---|---|---|---|
| WT control group | 0.920 ± 0.036 | 0.819 ± 0.035 | 3.984 ± 1.057 | 7.682 ± 0.310 | 0.335 ± 0.090 |
| WT model group | 0.686 ± 0.188# | 0.712 ± 0.102# | 2.605 ± 0.729# | 6.812 ± 0.523# | 0.225 ± 0.137# |
| WT AITC group | 0.816 ± 0.160△ | 0.767 ± 0.065△ | 3.032 ± 0.961△ | 7.232 ± 0.453△ | 0.257 ± 0.167△ |
| WT NAC group | 0.857 ± 0.161△ | 0.803 ± 0.037△ | 3.519 ± 0.964△ | 7.441 ± 0.216△ | 0.278 ± 0.225△ |
| Nrf2-/- control group | 0.931 ± 0.049 | 0.805 ± 0.028 | 3.24 ± 1.148 | 7.962 ± 0.252 | 0.324 ± 0.016 |
| Nrf2-/- model group | 0.625 ± 0.242▽* | 0.634 ± 0.048▽* | 1.886 ± 0.822▽* | 6.204 ± 0.311▽* | 0.166 ± 0.015▽* |
| Nrf2-/- AITC group | 0.696 ± 0.321※ | 0.645 ± 0.096 | 1.903 ± 1.302 | 6.650 ± 0.204※ | 0.169 ± 0.102 |
| Nrf2-/- NAC group | 0.704 ± 0.146※ | 0.680 ± 0.054※ | 2.355 ± 0.883※ | 7.052 ± 0.344※ | 0.213 ± 0.156※ |
Data are presented as the mean ± standard deviation (n = 6). FEV, forced expiratory volume; FVC, forced vital capacity; PEF, peak expiratory flow; FEF, forced expiratory flow; Cdyn, Cldyn; WT, wild type; AITC, allyl isothiocyanate; NAC, N-acetylcysteine; Nrf2, nuclear factor erythroid 2-related factor 2. #p < 0.05 vs. WT control group, △p < 0.05 vs. WT model group, ▽p < 0.05 vs. Nrf2-/- control group, ※p < 0.05 vs. Nrf2-/- model group, *p < 0.05 vs. WT model group.
Nrf2 gene knockout can impair the efficacy of AITC-induced MRP1 expression
 | Fig. 2Effect of Nrf2 gene knockout on MRP1 expression.(A) Immunohistochemistry results of Nrf2, DJ-1, and MRP1 expression in different groups. (B) Nrf2, DJ-1 and MRP1 relative optical density results in the WT groups. #p < 0.05 vs. control group, *p < 0.05 vs. model group (n = 5). (C) DJ-1 and MRP1 relative optical density results in the Nrf2-/- groups. #p < 0.05 vs. control group, *p < 0.05 vs. model group (n = 5). Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; WT, wild type; AITC, allyl isothiocyanate; NAC, N-acetylcysteine.
|
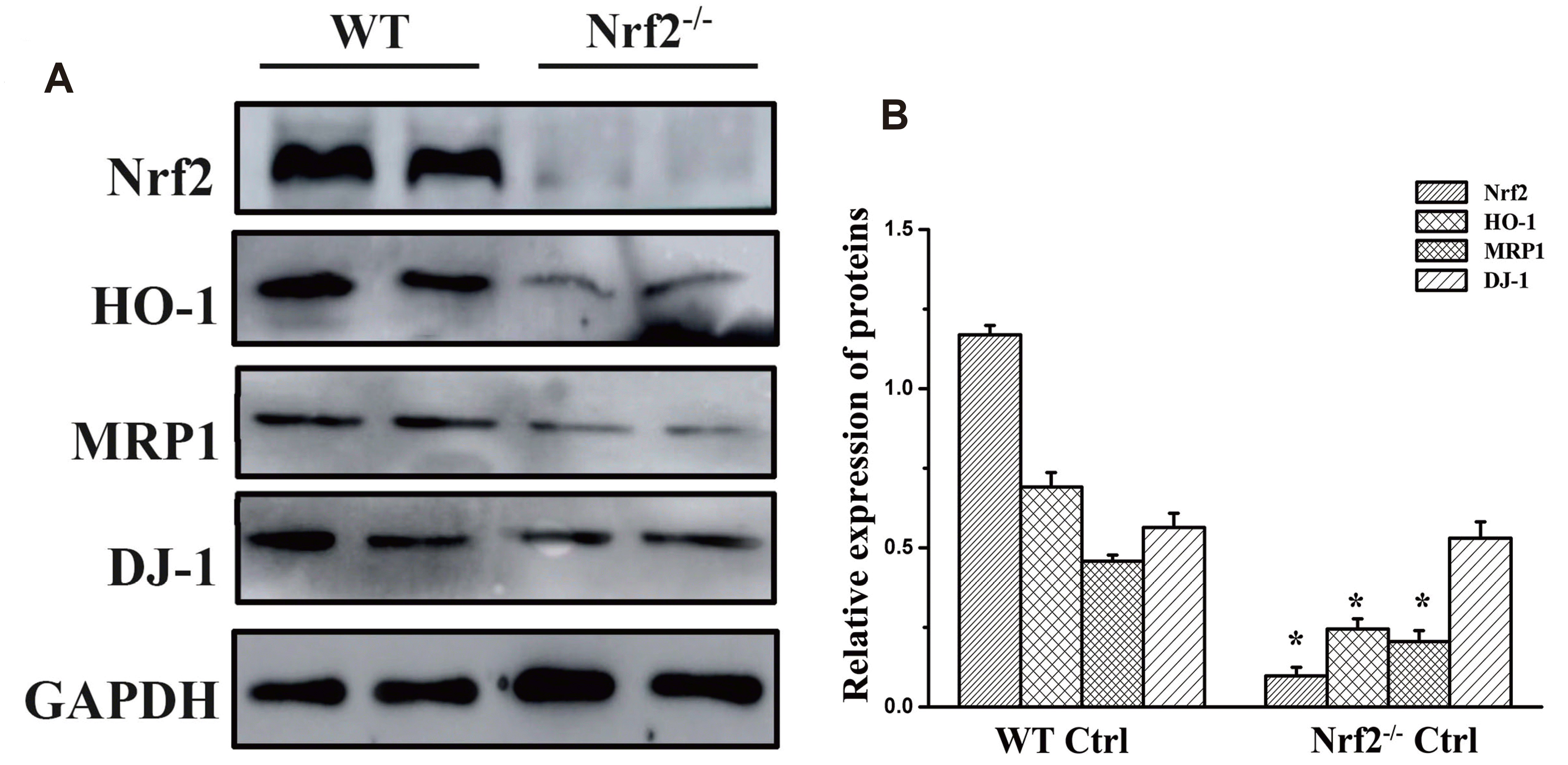 | Fig. 3Protein expression in the different groups.(A) Western blot results of Nrf2, DJ-1, MRP1, and HO-1 expression in the WT and Nrf2-/- control groups. GAPDH was used as the internal reference. (B) Quantitative analysis of the results of protein expression for each group. Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1; WT, wild type. *p < 0.05 vs. WT control group (n = 5).
|
 | Fig. 4Effect of AITC on MRP1 expression.(A) Western blot results of Nrf2, DJ-1, MRP1, and HO-1 expression in different groups. GAPDH was used as the internal reference. (B) Quantitative analysis of the results of protein expression in the WT groups. #p < 0.05 vs. control group, *p < 0.05 vs. model group (n = 5). (C) Quantitative analysis of the results of protein expression in the Nrf2-/- groups. #p < 0.05 vs. control group, *p < 0.05 vs. model group (n = 5). Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1; WT, wild type; AITC, allyl isothiocyanate; NAC, N-acetylcysteine.
|
 | Fig. 5Protein expression in 16HBE cells following CSE treatment.(A) MTT assay was used to determine the cellular toxicity of CSE in 16HBE cells. (B) Protein expression of Nrf2, DJ-1, MRP1, and HO-1 in 16HBE cells following treatment for different times with CSE. GAPDH was used as the internal reference. (C) Quantitative analysis of the results of protein expression in each group. CSE, cigarette smoke extract; 16HBE, human bronchial epithelial; Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1. *p < 0.05 vs. control group (n = 5).
|
 | Fig. 6Protein expression in 16HBE cells following treatment with AITC and CSE.(A) Protein expression of Nrf2, DJ-1, MRP1, and HO-1 in 16HBE cells following treatment with different concentrations of AITC. (B) Quantitative analysis of the results of protein expression in each group. *p < 0.05 vs. control CSE group (n = 5). (C) Protein expression of Nrf2, DJ-1, MRP1, and HO-1 in 16HBE cells following treatment with AITC and CSE. GAPDH was used as the internal reference. (D) Quantitative analysis of the results of protein expression in each group. #p < 0.05 vs. control group, *p < 0.05 vs. CSE group (n = 5). 16HBE, human bronchial epithelial; Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1; AITC, allyl isothiocyanate; CSE, cigarette smoke extract.
|
AITC has a positive effect on the expression of the Nrf2 positive regulator DJ-1
Silencing the DJ-1 gene accelerates CSE-mediated protein degradation
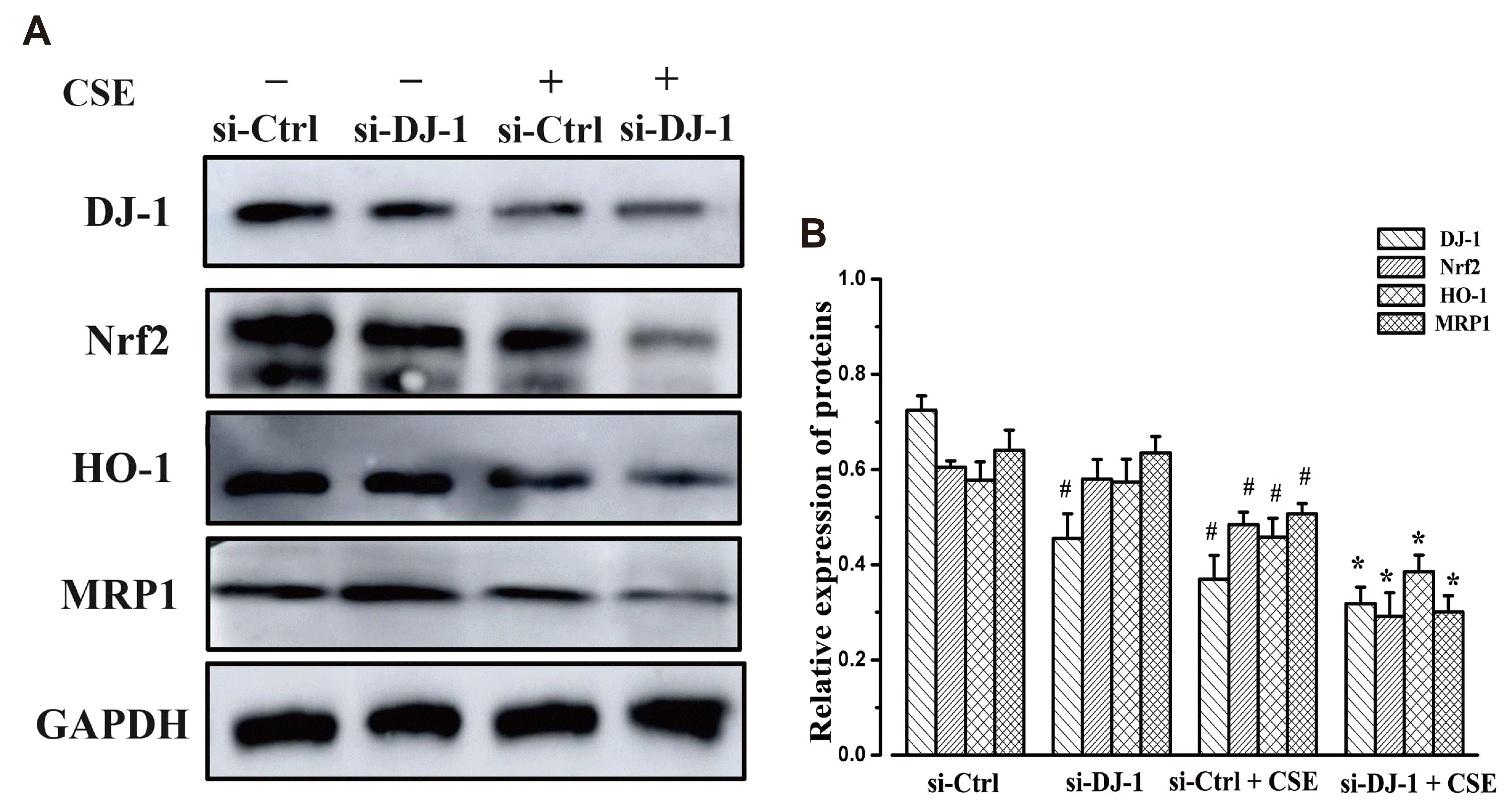 | Fig. 7Protein expression in 16HBE cells stimulated by CSE.(A) Protein expression of Nrf2, DJ-1, MRP1, and HO-1 in 16HBE cells following treatment with DJ-1siRNA/Lipofectamine 2000 complex and CSE. (B) Quantitative analysis of the results of protein expression in each group. 16HBE, human bronchial epithelial; Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1; CSE, cigarette smoke extract. #p < 0.05 vs. control group, *p < 0.05 vs. si-Ctrl+CSE group (n = 5).
|
AITC may upregulate the expression the of MRP1 via activating the DJ-1/Nrf2 axis
 | Fig. 8Effect of DJ-1 on MRP1 and AITC-induced Nrf2 and MRP1 expression.(A) Protein expression of Nrf2, DJ-1, MRP1, and HO-1 in 16HBE cells following treatment with DJ-1siRNA/Lipofectamine 2000 complex and AITC. (B) Quantitative analysis of the results of protein expression in each group. #p < 0.05 vs. control group, *p < 0.05 vs. si-Ctrl+AITC group (n = 5). (C) mRNA expression of Nrf2, DJ-1, MRP1 and HO-1 in 16HBE cells following treatment with DJ-1siRNA/ Lipofectamine 2000 complex and AITC. GAPDH was used as the internal reference. #p < 0.05 vs. control group, *p < 0.05 vs. si-Ctrl+AITC group (n = 6). 16HBE, human bronchial epithelial; Nrf2, nuclear factor erythroid 2-related factor 2; DJ-1, PARK7; MRP1, multidrug resistance-associated protein 1; HO-1, heme oxygenase-1; AITC, allyl isothiocyanate.
|




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download