INTRODUCTION
METHODS
Cells
Reagents and antibodies
Plasmids
Transient transfection and reporter assays
Western blot analysis
Statistical analysis
RESULTS
Differential effect of PML on OSM-induced STAT-3 transcriptional activity
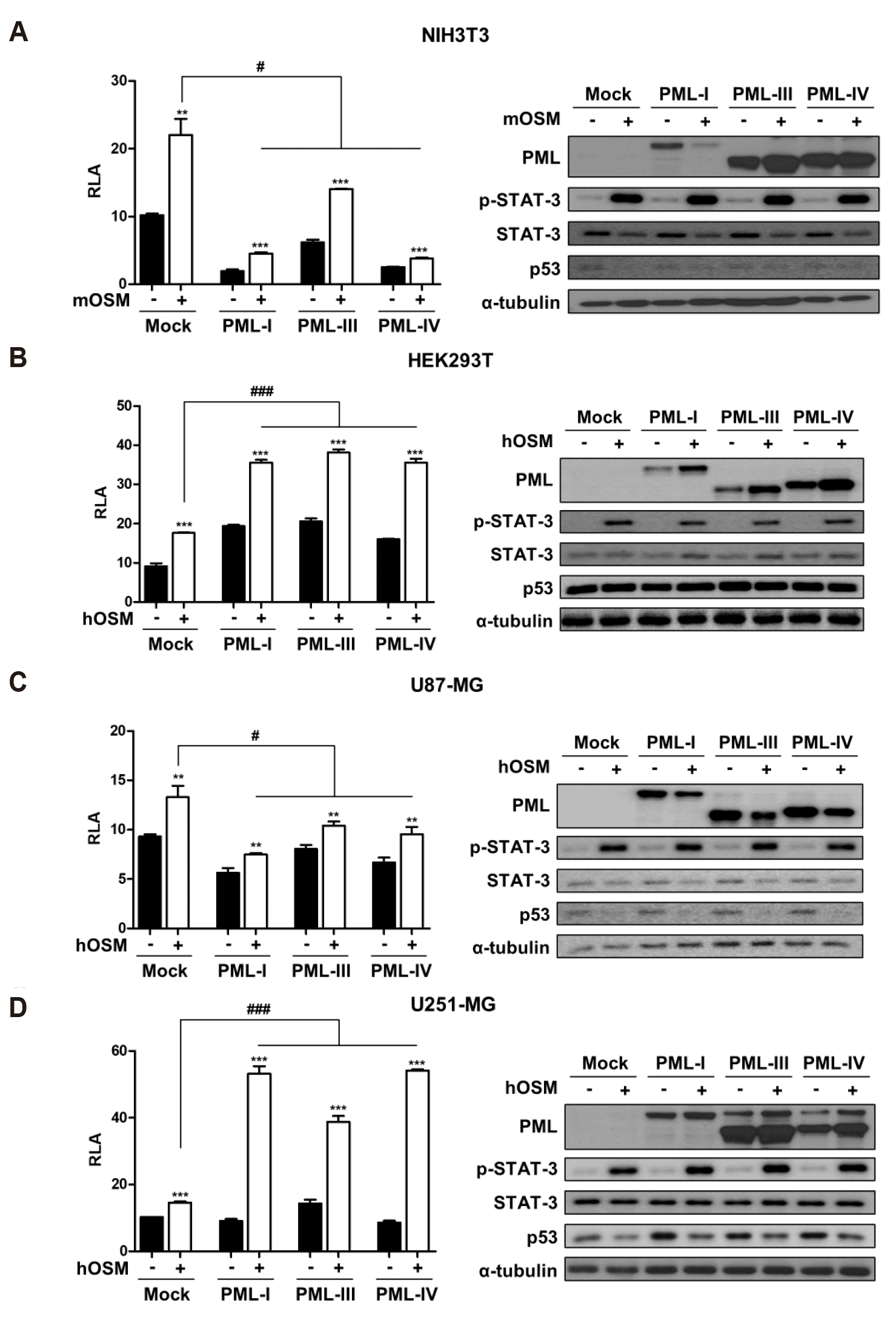 | Fig. 1PML affects differently OSM-induced STAT-3 transcriptional activity in immortalized cells and glioma cells.(A) NIH3T3, (B) HEK293T, (C) U87-MG, and (D) U251-MG cells were co-transfected with the STAT-3-Luc reporter and either empty vector or PML isoform expression vectors. At 24 h after transfection, cells were either untreated or treated with OSM (10 ng/ml) for 24 h and then assayed for luciferase activity. The pCMV-β-galactosidase vector was included to normalize transfection efficiency. Data are presented as fold increase in relative luciferase activity (RLA) compared with RLA in the absence of OSM. PML protein expression and OSM-induced STAT-3 phosphorylation were verified for each assay by immunoblotting. The results are representative of three independent experiments. mOSM, murine OSM; hOSM, human OSM. **p < 0.01, ***p < 0.001 vs. untreated control; #p < 0.05, ###p < 0.001 between cells were transfected with PML expression vectors and cells were transfected with mock vector in the presence of OSM.
|
Detection of endogenously expressed PML and wild-type and mutant p53
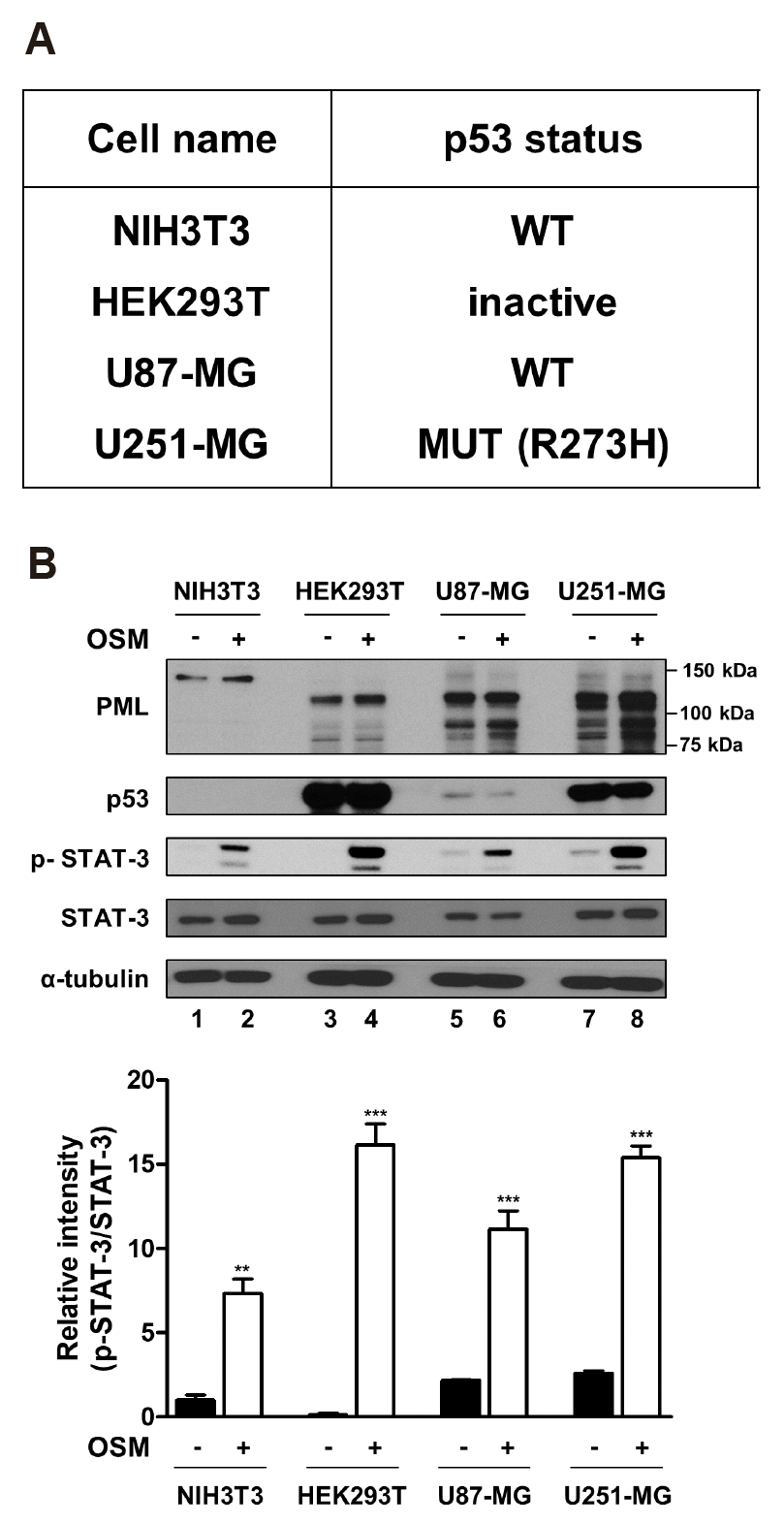 | Fig. 2The status of p53 and differential protein expression level.(A) The table indicates the p53 status of cell lines used in this study. p53 status was determined through the TP53 web site (http://p53.fr). (B, upper) NIH3T3, HEK293T, U87-MG, and U251-MG cells were either untreated or treated with OSM (10 ng/ml) for 24 h, and the lysed in RIPA buffer. Whole cell lysates were analyzed by immunoblotting using anti-PML, anti-p53, anti-p-STAT-3, anti-STAT-3, anti-α-tubulin antibodies. (B, lower) The graph represents the normalized intensities of p-STAT-3 against those of STAT-3 determined from three independent experiments. WT, wild-type. **p < 0.01, ***p < 0.001 vs. untreated control.
|
Opposite effects of PML-IV on OSM-mediated STAT-3 activity, depending on the p53 status
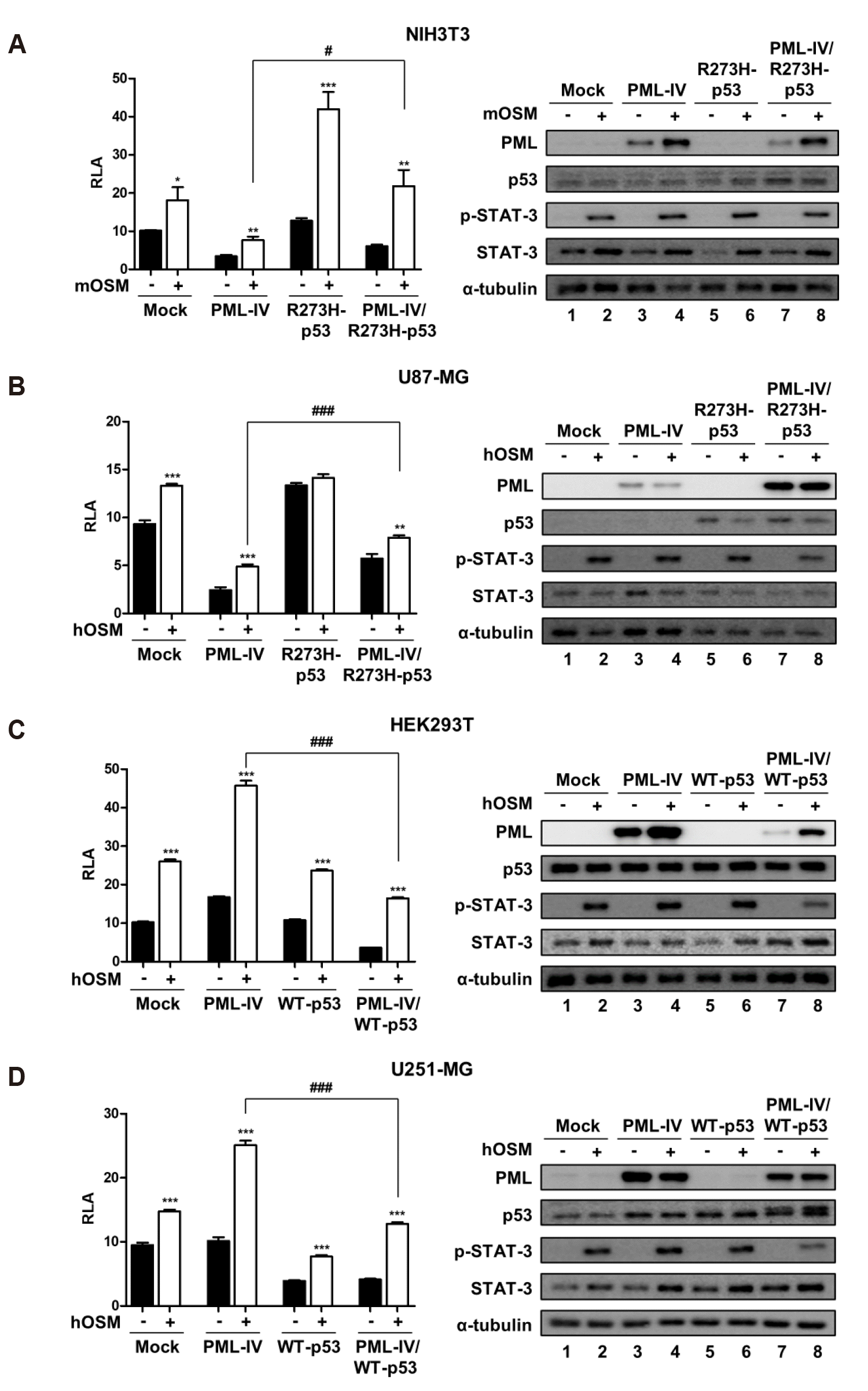 | Fig. 3The opposite effects of PML-IV on OSM-induced STAT-3 activity in p53-status dependent manner.(A) NIH3T3 and (B) U87-MG cells were transfected with either PML-IV or R273H-p53 or combination PML-IV and R273H-p53. (C) HEK293T and (D) U251-MG cells were transfected with either PML-IV or WT-p53 or combination PML-IV and WT-p53. At 24 h after transfection, cells were either untreated or treated with OSM (10 ng/ml) for 24 h and then assayed for luciferase activity. Data are presented as fold increase in relative luciferase activity (RLA) compared with RLA in the absence of OSM. PML and p53 protein expression and OSM-induced STAT-3 phosphorylation were verified for each assay by immunoblotting. The results are representative of three independent experiments. mOSM, murine OSM; hOSM, human OSM; WT, wild-type. *p < 0.05, **p < 0.01, ***p < 0.001 vs. untreated control; #p < 0.05, ###p < 0.001 between cells were transfected with PML-IV and cells were transfected with either combination PML-IV and R273H-p53 mutant or combination PML-IV and WT-p53 in the presence of OSM.
|
The effects of PML-IV and its deletion constructs on OSM-induced STAT-3 activity
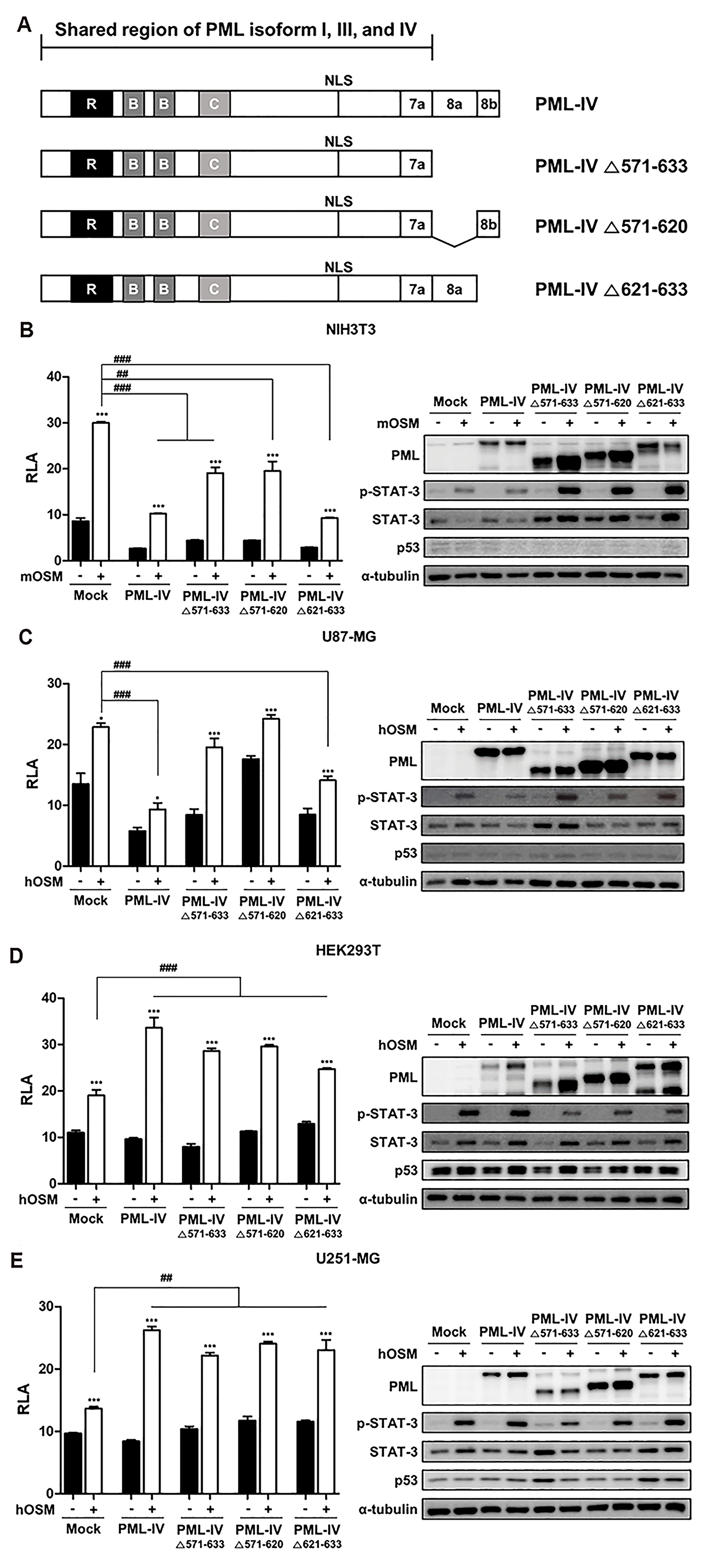 | Fig. 4The effects of PML-IV and its deletion constructs on OSM-mediated STAT-3 activity.(A) Depiction of the PML-IV deletion constructs generated in this study. (B) NIH3T3, (C) U87-MG, (D) HEK293T, (E) U251-MG cells were co-transfected with the STAT-3-Luc reporter and empty vector, PML-IV, and PML-IV deletion plasmids. At 24 h after transfection, cells were either untreated or treated with OSM (10 ng/ml) for 24 h and then assayed for luciferase activity. The pCMV-β-galactosidase vector was included to normalize transfection efficiency. Data are presented as fold increase in relative luciferase activity (RLA) compared with RLA in the absence of OSM. PML protein expression and OSM-induced STAT-3 phosphorylation were verified for each assay by immunoblotting. The results are representative of three independent experiments. mOSM, murine OSM; hOSM, human OSM. *p < 0.05, ***p < 0.001 vs. untreated control; ##p < 0.01. ###p < 0.001 between cells were transfected with either PML-IV or PML-IV deletion vectors and cells were transfected with mock vector in the presence of OSM.
|




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download