I. Introduction
Health data constitute a significant source of medical information and can be used after suitable processing for diagnosis and treatment. They comprise a huge number of records, which may conceal significant patterns and dependencies. The vast amounts of data generated as well as the increased complexity of the health field make it hard to extract hidden information. A data warehouse is a proper solution for data integration because it permits the management and querying of data in various ways and forms. Retrieving data from multi-dimensional data warehouse supports the revelation of hidden and important information. Thus, applying data mining techniques to data warehouses that store healthcare data for knowledge discovery is appropriate because such data is already cleaned, grouped, and stored [
1]. Thus, strategic decisions can be made to assist healthcare organizations.
Knowledge and experience are essential in doctors’ decision-making process. If decisions are made without proper caution, undesirable results can be encountered in the medical field, where the error tolerance is very low [
2]. The use of data warehousing and data mining techniques for decision support has emerged as a new direction in the healthcare field. Decision support systems built using data warehousing and data mining techniques are information system applications that assist healthcare professionals in making the best decisions for patients by providing the most up-to-date information.
Cancer is a complicated disease that has many different types of variables and fluctuates rapidly. Additionally, there are many factors that should not be disregarded during diagnosis, follow-up, and treatment [
1]. With the use of the decision support system based on data warehousing and data mining for cases of cancer, it will be possible to shorten the time needed for decision-making by healthcare providers. This will enable them to offer improved quality of care and achieve more positive outcomes.
According to the Global Cancer Observatory [
3], a total of 18.1 million new cases of cancer occurred globally during 2018, and there were 9.5 million cancer-related deaths. The most frequently diagnosed cancers were lung (11.6%), breast (11.6%), and colorectal (10.2%), while cancer deaths were mostly due to lung (18.4%), colorectal (9.2%), and stomach (8.2%) cancers. If this rate continues to grow, it is estimated that 19.3 million of people will have cancer by 2025 globally.
Cancer-related research is extensive, expensive, and complex, and it involves various healthcare organizations. In cancer research, data analysis of cancer incidence is conducted by age, gender, region, ethnicity, as well as economic and social factors that contribute to the assessment of population health needs. The analysis of cancer data warehouses using data mining techniques may discover hidden relations among patients’ data, cancer treatment, and disease surveillance [
4]. It is important to note that cancer care services are intrinsically multi-disciplinary, involving primary care physicians, pathologists, oncologists, and surgeons [
5]. The efficacy of these services will be increasingly affected by the shortage of oncology professionals and the limited number of specialists in complex scientific disciplines that involve data science, cancer treatment, and surveillance [
6].
In this study, we developed a clinical data warehouse for lung and ovarian cancer data provided by the United States National Cancer Institute (NCI;
https://www.cancer.gov). These data include demographic information of the patient, past medical history, general health status, treatment modalities, and cancer characteristics [
7]. The main contribution of this research is that for the first time the constellation schema is used to combine data from different cancer types to extract useful information related to hidden associations among medical features by data querying and analysis. The fact constellation schema consists of multiple fact tables, which share dimension tables. In fact, the fact constellation schema consists of more than one-star schema at a time by providing a flexible schema in which complex queries can be used to access data from the data warehouse. Other cancer data warehouse studies have been reported in the literature; however, they have concentrated on a specific type of cancer [
8–
13]. Our proposed system is designed to include data from both lung and ovarian cancer to find some possible hidden correlations among attributes. The aim of this study was to develop a clinical data warehouse for lung and ovarian cancer to be further used in data mining methods and decision support systems.
There are data warehousing, data mining, and decision support system implementations for healthcare information systems worldwide. Gorgionne et al. [
10] discussed how data warehousing, data mining, and decision support systems can reduce the national cancer hardship related to pharyngeal cancers. Their proposed system evaluates the efficacy of specified treatments and interventions with the formulations, detects cancer patterns in general and special populations, organizes relevant claims data, and formulates models that explain the patterns.
Abidi and Abidi [
14] introduced an integrated clinical evidence system designed to enhance clinical evidence with technology-mediated clinical evidence. Wu et al. [
15] demonstrated that the integration of clinical decision support into computer-based patient records can enhance patient safety, reduce medical errors, improve patient outcomes, and decrease unwanted practice variations.
Bellaachia and Guven [
9] used data mining techniques to predict the survivability rate of breast cancer patients. They implemented back-propagated neural network, naive Bayes and C4.5 decision tree algorithms. They found that the C4.5 algorithm’s performance was much better than that of the other two techniques. Wah and Sim [
12] developed a clinical data warehouse for lymphoma to improve the quality of diagnosis and treatment recommendation decision-making.
Sheta and Eldeen [
16] evaluated the architecture of a healthcare data warehouse specific to cancer diseases. They built the cancer data warehouse to integrate an operational database and medical files. Analysis of the data is facilitated by using OLAP (online analytical processing) cubes and viewing multilevel details of the data. According to the model developed in [
11], the naive Bayes method is the most effective way to predict patients with lung cancer. Arous et al. [
8] integrated two different operational health systems that include pancreatic cancer data. They explained the challenges they faced because the data types in the databases differed. Ramachandran et al. [
17] developed a clinical data warehouse to identify potential cancer patients, which uses classification, clustering, and prediction data mining technologies. After the preprocessing step, the data is clustered using a K-means clustering algorithm. Their research can help detect a person’s predisposition to develop cancer before going for clinical and lab tests.
The remainder of this paper is organized as follows. Section II describes the methods applied related to the data warehouse domain and the research procedures used. Section III presents the results and describes the implementation of the proposed clinical data warehouse. Finally, Section IV provides our interpretation of the results, concluding remarks, and future research plan.
Go to :

II. Methods
In this study, a clinical data warehouse for both lung and ovarian cancer data was developed for data mining and clinical decision support. The data required for the study were obtained from the NCI’s prostate, lung, colorectal, and ovarian cancer screening trial. Three major tasks had to be performed to prepare the data, namely, data acquisition, storage, and information delivery as depicted in
Figure 1 [
16].
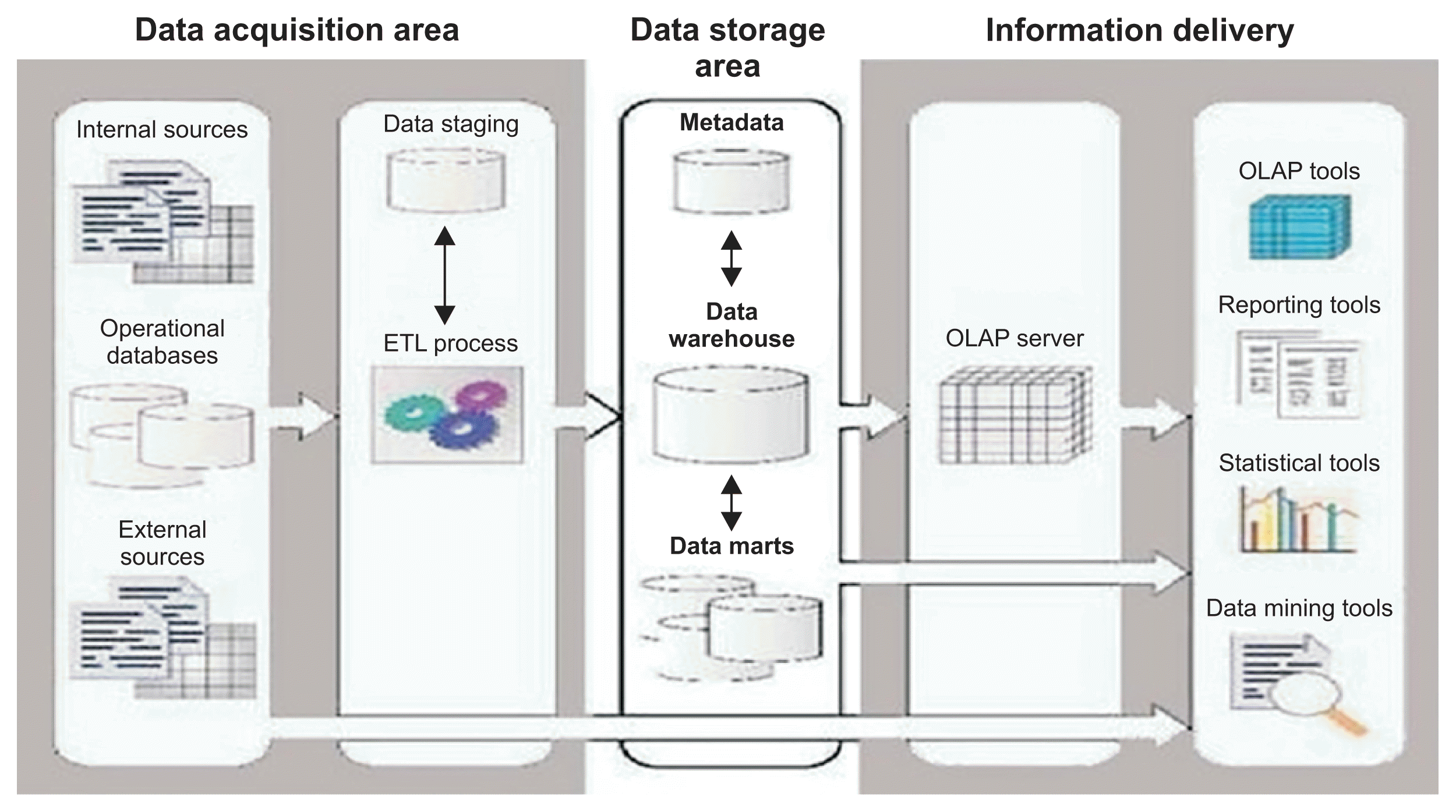 | Figure 1Proposed project architecture. Adapted from Sheta and Eldeen [ 16]. 
|
Pre-processing of data is an important phase in the implementation of data warehouse applications. For data quality control, it is vital to pre-process the data to ensure that it is clean and meaningful. Lung and ovarian cancer data were cleaned to remove mistakes, repetitions, and stored in the data warehouse as a first stage. It was ensured that the data warehouse would conform to the subject-oriented, integrated, time-variant and non-volatile conditions of the data warehouse approach presented in [
18].
1. Data Warehouse
A data warehouse schema is composed of fact and dimension tables. The fact table is connected to a number of dimension tables with many-to-one relationships. The term “fact” represents a business measure. A fact table includes two kinds of columns, that is, two or more foreign keys that connect primary keys and numeric attributes (known as measures) on which calculations can be made.
Two of the most popular and well-known schemas for implementing a multi-dimensional model are the star schema and the snowflake schema [
19]. The star schema adopts the relational model for the representation of multi-dimensional data consisting of a fact table and a number of non-normalized dimension tables.
Figure 2 presents an example of a star schema for medical records. Each tuple of the fact table stores data from a patient with one diagnosis having a specific therapy type applied on one day. Other possible granularities for the time dimension include week, month, and year. The granularity decision influences the degree of detail for the recorded data and the size of the database.
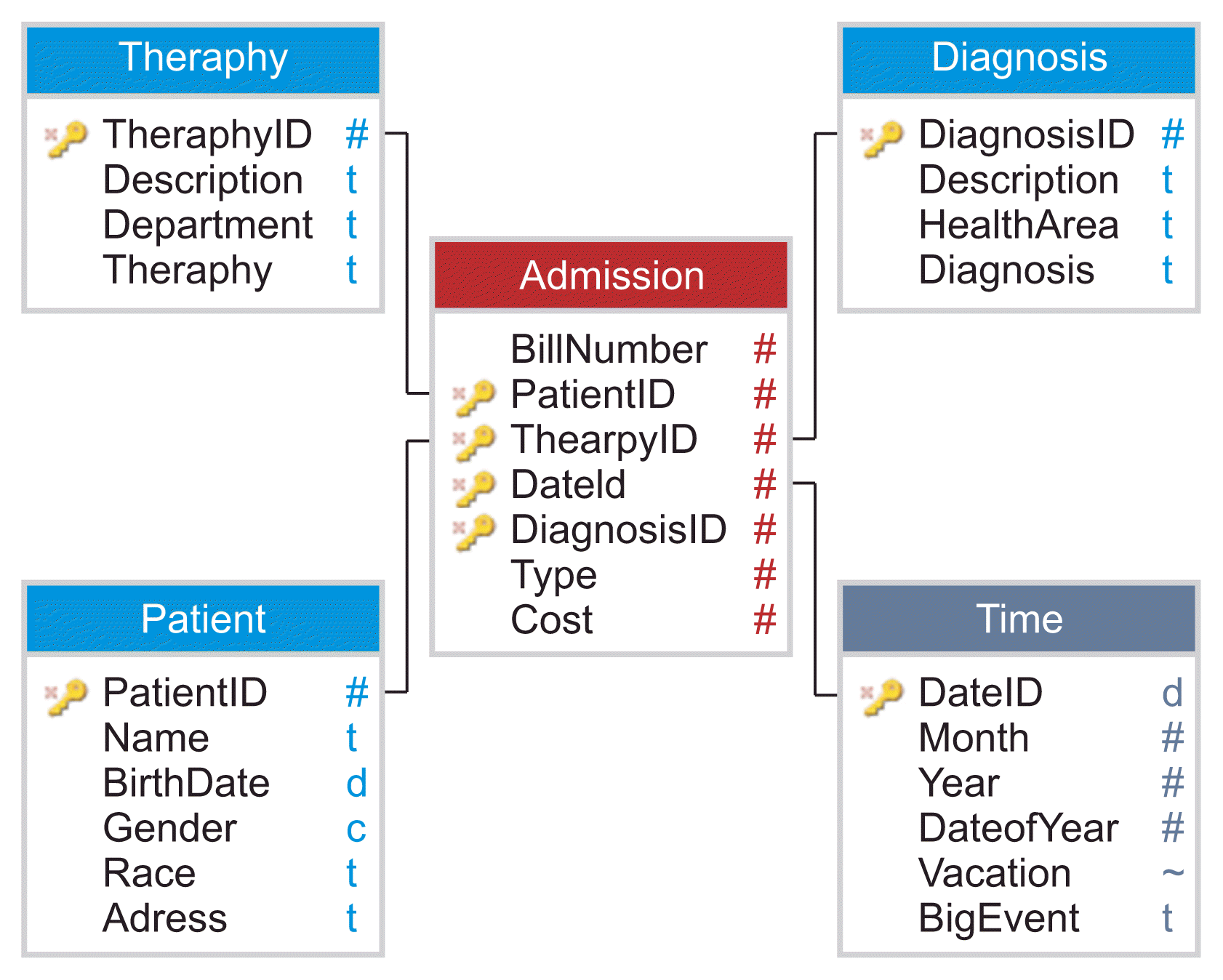 | Figure 2Star schema for medical records. 
|
Each dimension consists of one or more hierarchies and represents a semantically related concept within the modeled domain. Dimensional attributes are descriptive, textual values for describing the dimensional value. Dimension tables have fewer rows than fact tables. Each dimension is defined by a single primary key that serves as the basis for the referential integrity with any given fact table to which it is joined [
19].
A snowflake schema is obtained from a star schema, and it is made up of a fact table and many dimensions related to that fact. In this type of schema, dimension data are grouped into multiple (totally or partially) normalized tables instead of one large non-normalized table. An example of a patient admission snowflake schema is shown in
Figure 3. With snowflake modeling, the number of the tables and the required joins increase.
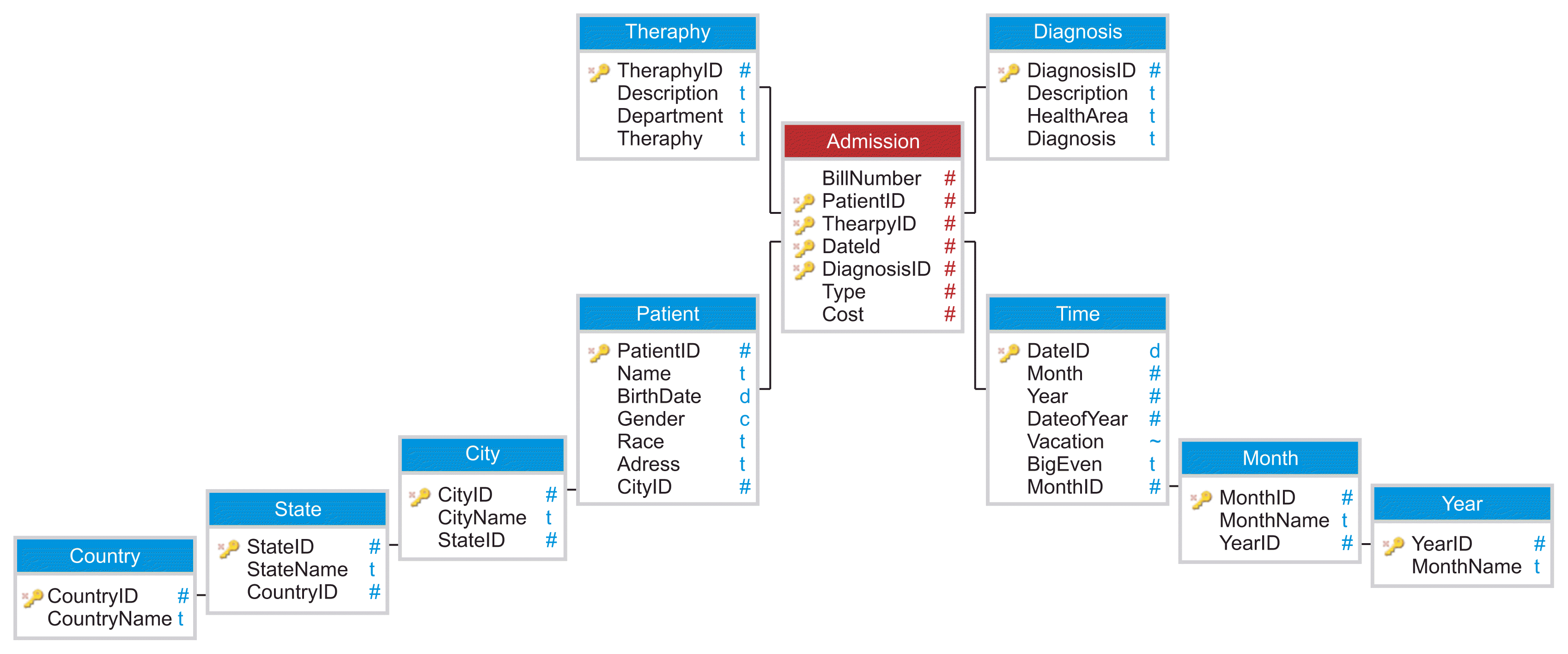 | Figure 3Snowflake schema for medical records. 
|
A fact constellation schema consists of a collection of several fact tables. They share dimension tables, which are called conformed dimensions. It can be considered as an extension of a star schema. Although it is more complicated than the star and snowflake schemas, it is generally utilized. For designing the lung and ovarian cancer diseases data warehouse, the fact constellation schema model was adopted as a multi-dimensional data modelling approach because it can be used as a collection of numerous star schemas.
Go to :

III. Results
1. PLCO Data
The NCI, the main institution for cancer research in the United States, provided the data used for this research. The NCI manages the National Cancer Program, which conducts and supports research, training, health information dissemination, and other programs with respect to the cause, diagnosis, prevention, and treatment of cancer, rehabilitation from cancer, and the continuing care of cancer patients and the families of cancer patients [
7].
The prostate, lung, colorectal, and ovarian (PLCO) dataset was created through a large-scale, randomized study to determine whether certain screening tests will reduce the number of deaths from these cancers [
7]. Within the scope of this research, data from 3,594 lung and 3,914 ovarian cancer cases were studied. These data included demographic information about the patient, past medical history, general health status, smoking history, treatment modalities, and cancer characteristics. Some attribute examples from the dataset are presented in
Table 1.
Table 1
Attribute examples from the dataset
|
Attribute |
Description |
Text format |
|
agelevel |
Patient age level |
0 = “<=59” |
|
1 = “60–64” |
|
2 = “65–69” |
|
3 = “>=70” |
|
|
sqx_fh_lung |
Family history of lung cancer |
0 = “No” |
|
1 = “Yes” |
|
|
lung_stage_m |
M stage component (distant metastases) |
1 = “MX” |
|
2 = “M0” |
|
3 = “M1” |
|
99 = “Not available” |
|
|
curative_radl |
Had radiation treatment for lung cancer |
0 = “No” |
|
1 = “Yes” |
|
|
bronchit_f |
Did the participant ever have chronic bronchitis? |
0 = “No” |
|
1 = “Yes” |
|
|
Sqx_smk30days |
Smoke in the last 30 days |
1 = “Every day” |
|
2 = “Some days” |
|
3 = “Not at all” |

2. Clinical Data Warehousing Stages
Data integration is very important for healthcare organizations because these data must be provided as input to clinical decision support systems. Data from different operational systems are collected in a common repository and made available for operations such as querying and analysis using the data warehouse approach.
The fact constellation schema can support consistent, integrated, and flexible sources of data, so it was adopted in this research after thoughtful consideration of the available types of cancer data provided by the NCI.
To build the lung and ovarian cancer clinical data warehouse, preprocessing operations were applied to data. Data processing was the most important and time-consuming part of the process of designing the data warehouse, which included various processes, such as filtering, cleaning and transforming the data, to ensure better quality and accurate results. In this study, the data warehouse was developed with Microsoft SQL Server 2012. First, the data were extracted from the source system. SQL server integration services (SSIS) were used for the extract-transform-load (ETL) process. After the elimination of wrong and inconsistent data, the data were transformed and loaded into the data warehouse.
The data are stored in a fact constellation schema model using a multi-dimensional modeling approach to perform OLAP operations. The multi-dimensional data model is based on concepts such as cube, dimension, and hierarchy. The fact constellation schema model used is easy to understand and appropriate for query performance. As seen in
Figure 4, the fact table contains keys representing each of the dimension tables. The lung and ovarian cancer data warehouse model created using the Microsoft SQL Server has 14 dimension tables and 2 fact tables that contain the following: (1) complications, diagnoses, treatments, X-ray results, and X-ray anomalies for each cancer type, (2) patient information, diet history, dietary data and time dimensions shared by both fact tables, (3) one lung cancer fact table, and (4) one ovarian cancer fact table.
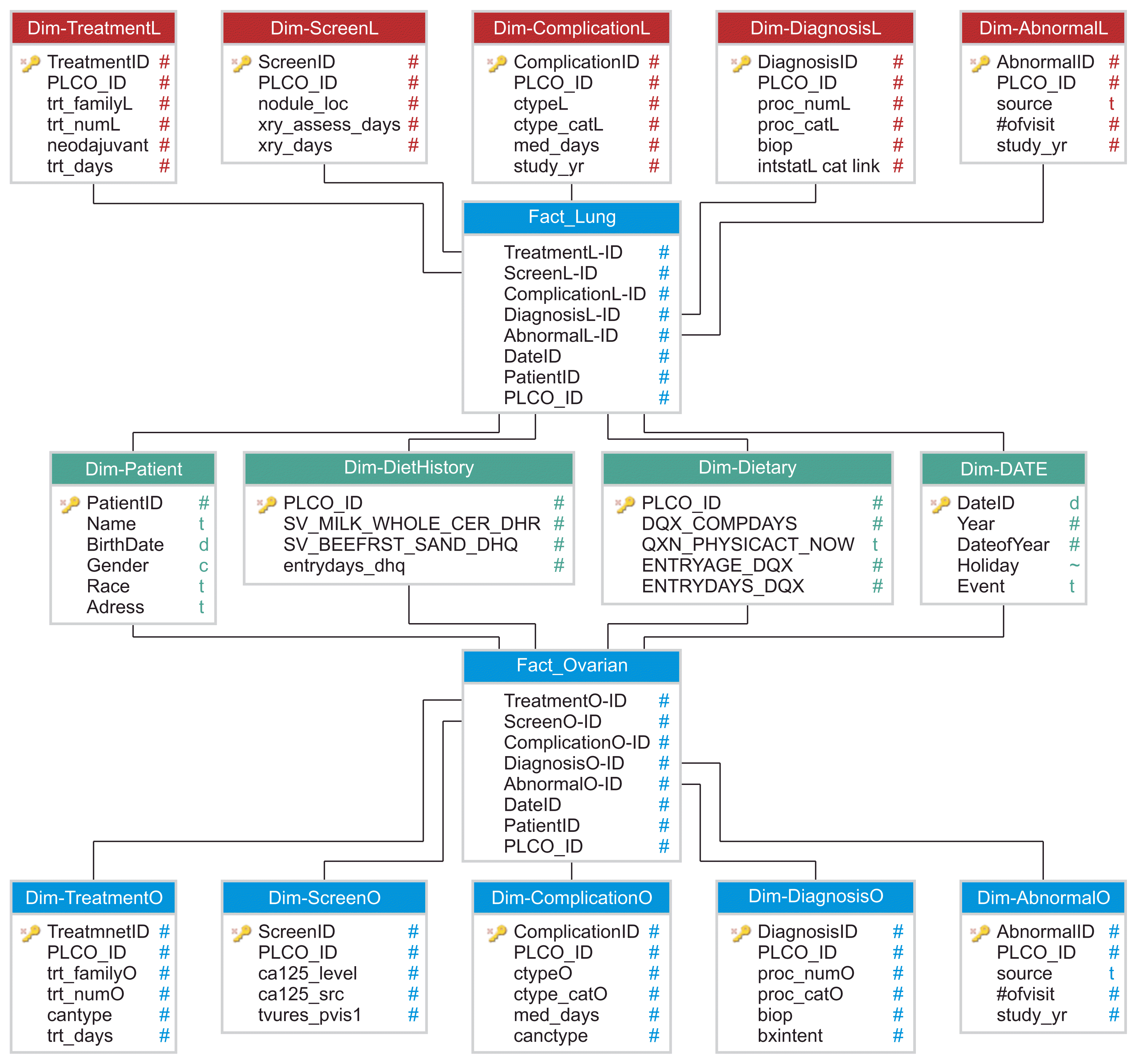 | Figure 4Lung and ovarian cancer clinical data warehouse fact constellation schema model. 
|
At a later stage, the SQL server analysis services (SSAS) were used to build cubes using the lung and ovarian clinical data warehouse as the data source. A single cube was built using all dimensions in the data warehouse. This is to allow analysis of the diet history factor with respect to each category of complications, abnormalities, location, age, sex, and time. Building cubes enables users to use OLAP tools for interactive analysis of multi-dimensional data at various granularity levels. Data cubes store summarized data that precomputed measures such as count() and sum() retrieve faster in terms of query processing times.
3. OLAP Queries
The established clinical data warehouse was designed to respond to large and complex queries. It is intended to help medical analysts and physicians in making decisions through diverse perspectives. Several sample OLAP queries are presented below for the lung and ovarian cancer clinical data warehouse.
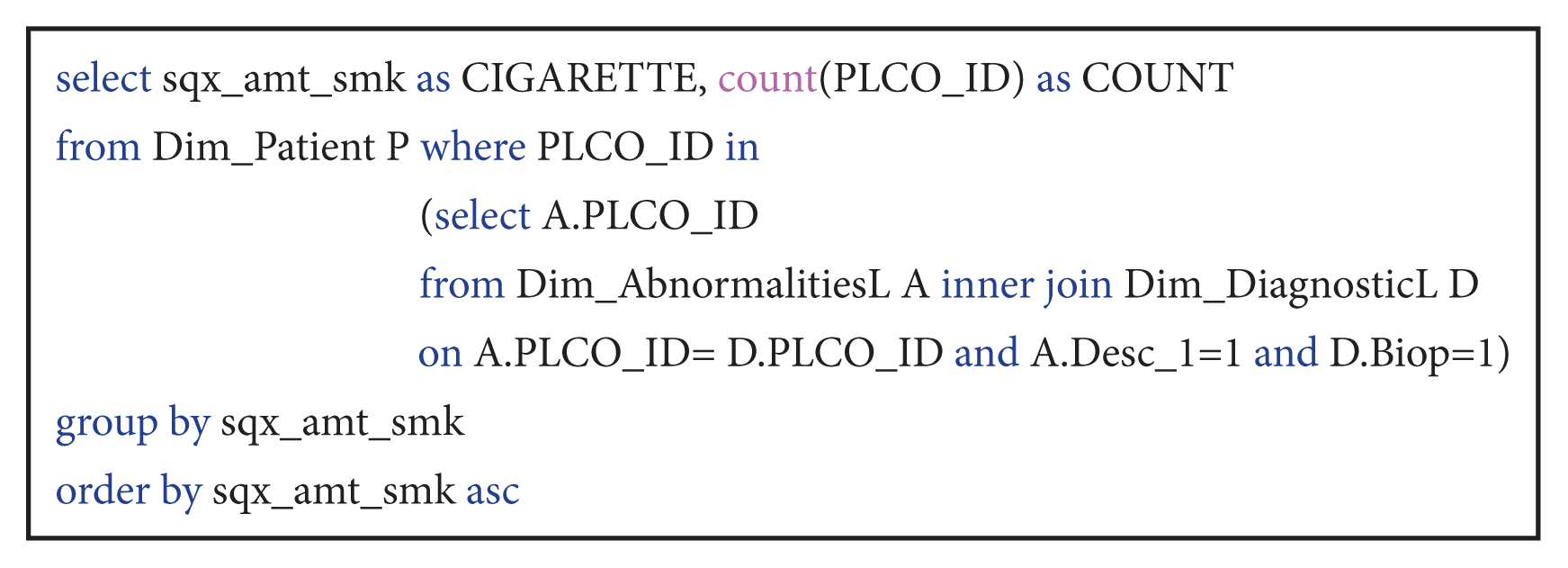 | Query 1How is the distribution of patients with nodules and biopsies according to the number of daily cigarettes? 
|
This query counted the number of patients grouped and ordered by the daily smoked cigarettes who received a biopsy and had nodules. The query is very important to prove the danger of smoking.
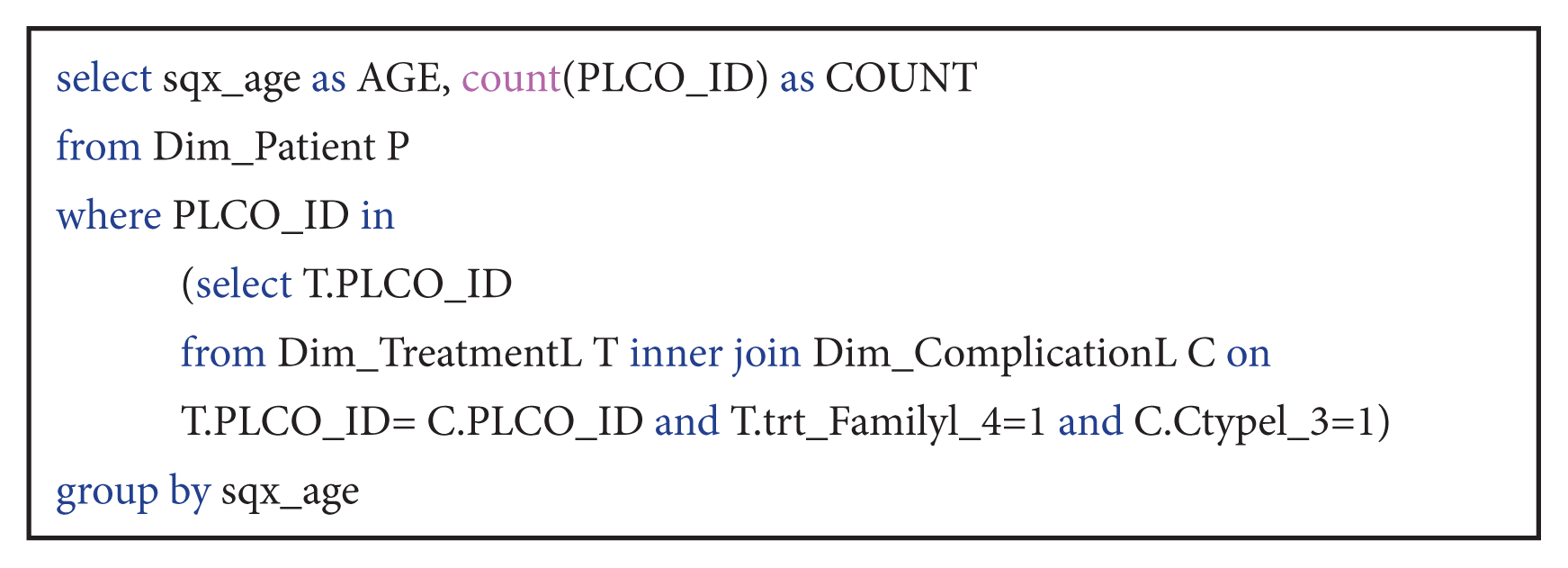 | Query 2What is the distribution of patients with complication Pnömotoraks, collection of air in the pleural cavity, and those treated with chemotherapy according to age? 
|
This query counted number of patients grouped by their age that had Pnömotoraks complication and were treated with chemotherapy. The result table helps doctors to evaluate specific age groups that have both conditions.
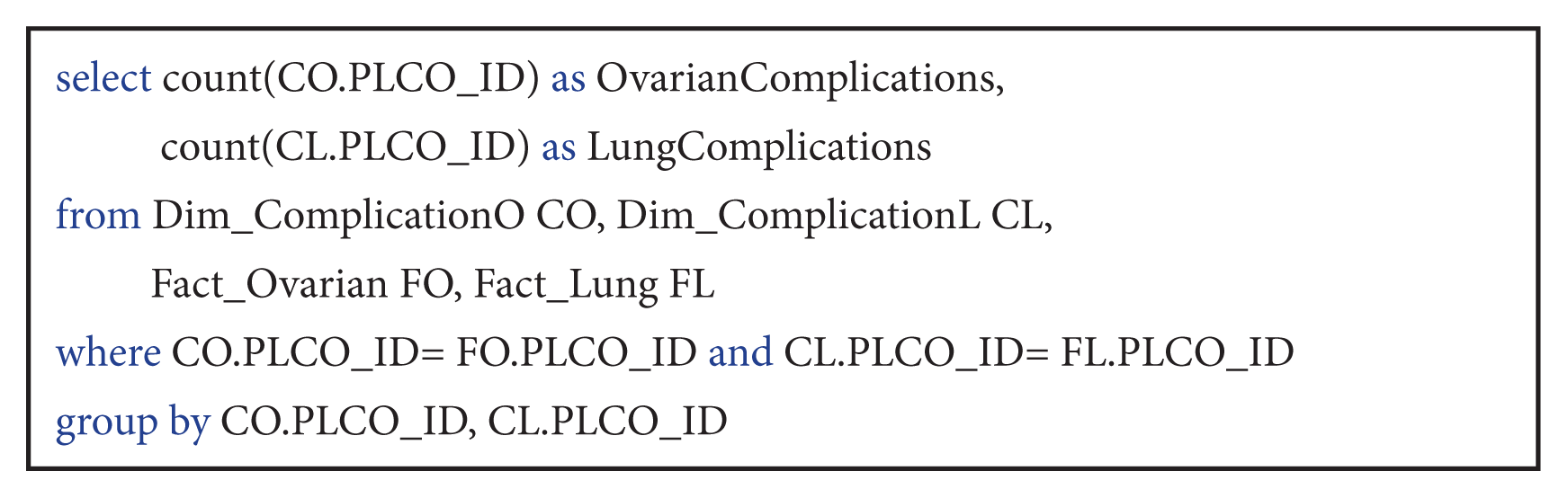 | Query 3Compare the number of complications in ovarian and lung cancer patients. 
|
This query combined data from both cancer types, lung and ovarian. Therefore, both fact tables from the fact constellation schema were used, Fact_Lung and Fact_Ovarian. The query is very important for statistical medical data analysis.
This query was used to extract patients’ personal data related to both types of cancers and specific treatment. In spite of advanced query complexity,
Query 4 is easily expressed using the fact constellation schema by performing four join operations.
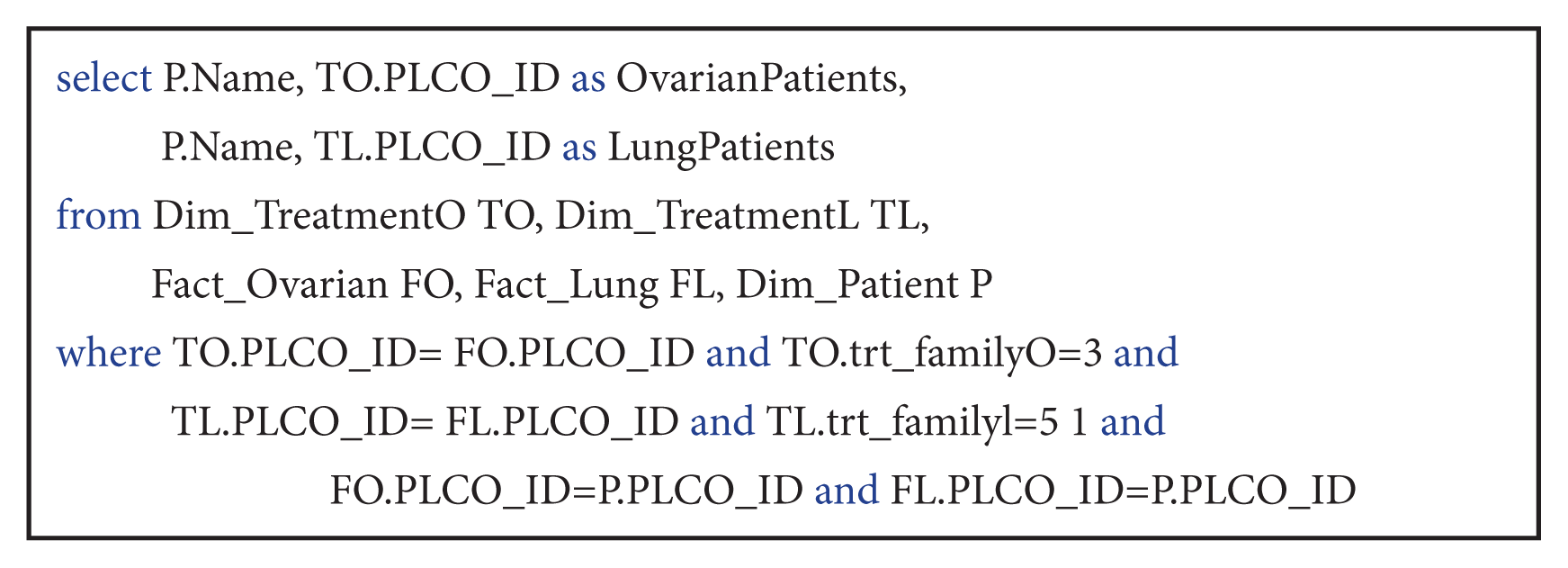 | Query 4List the PLCO_ID numbers and names of patients who received “non-curative” treatment for both lung and ovarian cancer. 
|
Go to :

IV. Discussion
Data warehouses are capable of integrating massive data from various sources for analysis and querying purposes. The healthcare field can benefit significantly from data warehousing for the study, prediction, and treatment of cancer, and in general, for the improvement of the quality of human lives.
In this study, a clinical data warehouse was designed for lung and ovarian cancers with data collected from the NCI. The main goal of this research was to analyze and store data regarding two different cancer types in one single data warehouse. We ensured that the data warehouse would conform to the subject-oriented, integrated, time-variant and nonvolatile conditions of the data warehouse approach presented by [
18]. Data were integrated through preprocessing, transformation, and selection operations. In the design of the data warehouse, the fact constellation schema model, a multi-dimensional data modeling approach, was used. This model succeeded in responding to complex queries, and the analysis of data was facilitated by using OLAP cubes and viewing multi-level data details.
Indisputably, the fact constellation schema is the most challenging data warehouse design architecture. For the first time, real medical data were used combining two different cancer types using fact constellation modelling to extract semantically useful information to improve the decision-making process for cancer patients. The present approach also provides the ability to access data in the data warehouse using complex queries because dimensional attributes are shared by a number of fact tables, unlike the star and snowflake schemas. The fact constellation schema is capable of dealing with complex systems because the relationships between fact and dimensional tables are more easily understood. The medical field requires advanced data analytical processing by studying real critical medical data and this work addresses this need.
We plan to integrate prostate and colorectal cancer data into the already built clinical data warehouse and then to develop a decision support system for adapting more data analysis and mining tools. Hence, we can analyze cancer diseases, find correlations among attributes and cost of treatment for these diseases, death rates for specific types of cancer, and the impact of particular drugs on each disease. We expect that the final implementation of the decision support system will greatly assist cancer informatics research.
Go to :








 PDF
PDF Citation
Citation Print
Print








 XML Download
XML Download