INTRODUCTION
Feline herpesvirus 1 (FHV-1) belongs to the genus Varicellovirus, subfamily Alphaherpesvirinae in the family Herpesviridae. The FHV-1 genome has approximately 135,800 nucleotides including unique long (UL) and unique short (US), and terminal and inverted repeat long segments, which contain 78 open reading frames and have a G+C content of approximately 45% (1, 2). FHV-1 has 23 virion-associated proteins and its envelope consists of 13 glycoproteins: UL53 (gK), UL49.5 (gN), UL44 (gC), UL43, UL27 (gB), UL22 (gH), UL20, UL10 (gM), UL1 (gL), US4 (gG), US6 (gD), US7 (gI), and US8 (gE) (2). Four viral proteins have been identified as potential virulence factors: gE, gC, thymidine kinase (TK, UL23), and serine/threonine protein kinase (US3) (3). Glycoprotein B (gB, UL 27), a vital component of the surface of FHV-1 that is known to induce a protective immune response, is used as an enzyme- linked immunosorbent assay antigen for detection of FHV-1 antibodies (4).
Clinical signs of FHV-1 infection in cats are fever, nasal and ocular discharge, conjunctivitis, keratitis, and pneumonia; sometimes, abortion and fetal death can occur (5). Following the reduction in maternal antibodies, 6–9-week-old cats are more likely to be infected with FHV-1. Infections in cats have been reported in many countries (6, 7, 8, 9). Transmission typically occurs via the oronasal route, following contact with ocular discharge from acutely infected cats (10). Similar to latent infection reported in other herpesviruses, trigeminal ganglion FHV-1 infection can be reactivated by strong stress and immunosuppression, resulting in infectious virus shedding (11).
The diagnosis of FHV-1 infection is based on virus isolation with cell cultures, pathological findings, the direct fluorescent antibody test, polymerase chain reaction (PCR), real-time PCR, and serological assays (4, 12, 13). Virus neutralizing antibody titers measured with the virus neutralization assay are used to monitor sero-epidemiological status or to evaluate the immunogenicity of vaccination (14). Among the various diagnostic methods, virus isolation assesses the efficacy of trial feline vaccines and is used to establish serological diagnostic assays. It is also useful when developing new vaccines for FHV-1. Because FHV-1 has a global distribution, many FHV-1 strains have been isolated from cats and tigers (6, 7, 15). Although FHV-1 infection was examined in cats at a Korean animal shelter in 2008 (9), there has been no report of the successful isolation of Korean FHV-1 or its genetic features.
In this study, we isolated two FHV-1 isolates from naturally infected cats by Crandell–Rees feline kidney (CRFK) cell culture. They were designated FHV191071 and FHV191072; notably, they were propagated and passaged five times in the cells. In addition, we examined the properties of both isolates and determined the nucleotide sequences of the TK and gB genes of both isolates to evaluate their genetic relationships. The results of this study increase our knowledge of the biological and molecular features of Korean FHV-1 isolates.
Go to : 
MATERIALS AND METHODS
Samples and preparation
In this study, 48 samples of lung, spleen, brain, kidney, liver, and feces were prepared from 12 cats that had been sent to the Animal and Plant Quarantine Agency (APQA) for disease diagnosis in 2019. One gram of each sample was placed in a mortar and ground with a small amount of sea sand; 9 mL of Dulbecco’s modified Eagle’s medium (DMEM) was added to make a 10% homogenate. The homogenates were centrifuged at 2,500 × g for 15 min to remove tissue debris; the supernatants were filtered through a 0.45-μm syringe filter to avoid bacterial infection.
Virus isolation, growth kinetics, and titration
The CRFK (CRFK, ATCC, CCL-94) cells used for virus isolation were cultured in DMEM supplemented with two antibiotics (penicillin and streptomycin) and one antimycotic (amphotericin B), as well as 10% fetal bovine serum (FBS). Confluent CRFK cells in 24-well plates were washed twice with phosphate-buffered saline (PBS, pH 7.2), then incubated with 200 μL of each filtered sample at 37°C for 1 h. Subsequently, the liquid in the 24-well plates was removed and 1 mL of fresh DMEM containing 3% FBS was added to each well. Cells were incubated in a 5% CO2 incubator at 37°C for 7 days. If the CRFK cells in the second passage did not show any cytopathic effects (CPE) after 7 days, the samples were considered negative for virus isolation. The supernatant in wells of cells with CPE was harvested and inoculated into newly prepared CRFK cells in a 6-well plate for the second passage. The growth kinetics of the FHV191071 and FHV191072 isolates were examined twice to determine the most rapid growth time. Briefly, CRFK cells grown in 25-cm2 flasks were inoculated with isolates containing 100 50% tissue culture infectious dose (TCID50/mL) and harvested at 12-h intervals for 84 h. After three continuous freeze–thaw processes, the viral titers of both isolates were determined based on the presence of typical CPE as follows: 100 μL of each virus diluted 10-fold was distributed in 96-well microplates; 100 μL of fresh medium containing 2 × 105 CRFK cells and 10% FBS was added to the wells of the microplate. CRFK cells were observed for 5 days post-inoculation for typical CPE. Viral titers calculated by the Spearman–Karber method were expressed as the TCID50/mL. To identify the optimal cells for viral propagation, both isolates were inoculated into six cell types in 25-cm2 flasks: CRFK, Fcwf-4 (ATCC, CRL2728), BHK-21 (ATCC, CCL-10), A72 (ATCC, CRL-1542), Madin–Darby canine kidney (MDCK; ATCC, CCL-34), and Vero (ATCC, CCL81). After incubation for 5 days, each flask was frozen and thawed three times. Viral titers of the centrifuged solutions were measured using the method described above.
Immunofluorescent assay
CRFK cells infected with isolates in 96-well microplates were fixed with cold 80% acetone at –20°C for 15 min. The cold acetone was then discarded and the cells were reacted with a mouse anti-feline herpesvirus type 1 antibody (Bio-Rad, Hercules, CA, USA) at 37°C for 1 h; they were then stained with fluorescent isothiocyanate-conjugated goat-anti-mouse IgG + IgM antibodies (KPL Laboratories, Gettysburg, PA, USA) diluted 200-fold in PBS under the conditions described above. Subsequently, the cells were washed with PBS twice, then air-dried and examined under a 200× fluorescence microscope (TE2000-U, Nikon Instruments, Tokyo, Japan). Specific fluorescence of CRFK cells with CPE was considered to indicate FHV-1 infection.
Electron microscopy (EM)
Isolates were propagated in CRFK cells, then concentrated and purified for photography of intact viral particles as follows: CRFK cells infected with the FHV191071 and FHV191072 isolates were harvested at 48 h after inoculation, in accordance with the growth kinetics, then frozen and thawed three times. Polyethylene glycol-precipitated pellets were dissolved in TNE buffer (100 mM Tris-Cl, 100 mM NaCl, and 1 mM EDTA, pH 7.6) at 5% of the original volume. The concentrated samples were then subjected to sucrose purification, placed on Formvar-coated grids, and stained negatively with 1% uranyl acetate. To identify viral particles in infected cells, CRFK cells were prepared and inoculated with each isolate. At 30 h post-inoculation, cells were harvested and fixed with 2.5% glutaraldehyde and 1% osmium tetroxide solution in PBS. The fixed cells were embedded in resin and cut thinly with a microtome for staining; sliced cells were stained with uranyl acetate and lead citrate. Virus particles in the two isolates on the grid and sliced sections were examined under a Hitachi 7100 electron microscope (Tokyo, Japan).
Hemagglutination (HA) assay
For the HA assay, CRFK cells infected with the isolates were serially diluted two-fold with 50 μL PBS (pH 7.2); 50 μL of 0.5% erythrocytes obtained from cats, guinea pigs, and mice were then added and incubated at 4, 22, and 37°C for 1 h. The HA titers were expressed as the reciprocal of the highest dilution of FHV-1 that demonstrated an HA reaction.
PCR, sequencing, and phylogenetic analysis
DNA was extracted from both isolates using a DNA extraction kit (Bioneer, Daejeon, Korea), in accordance with the manufacturer’s instructions. DNA attached to the column was eluted with 50 μL of elution buffer provided in the kit. PCR was performed for diagnosis and cloning. Table 1 lists the primers for detection of FHV-1 and amplification of the full TK and gB genes. The reaction mixture for One-Step PCR kits (Bioneer) consisted of 5 μL denatured DNA, 1 μL each primer (50 pmol/μL), and 43 μL distilled water. A thermal cycler (Analytik Jena, Jena, Germany) was used to perform 40 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 45 s, with a final 10-min extension at 72°C. Approximately 5 μL of each PCR product was loaded on a 2.0% agarose gel and subjected to electrophoresis for 30 min.
Table 1.
Primers used for polymerase chain reaction analysis to amplify the thymidine kinase and glycoprotein B genes of the two isolates
Five PCR products of partial TK and gB genes for cloning were purified by gel extraction; each product was ligated into the pGEM®-T Easy vector system (Promega, Madison, WI, USA), in accordance with the manufacturer’s instructions. Selected white colonies grew in Luria-Bertani broth containing 50 μg/μL ampicillin. Plasmid DNA was extracted from several white Escherichia coli (DH5α) colonies; each DNA insert in the plasmid was identified by EcoR1 digestion (Bioneer). Sequencing was performed by Macrogen (Daejeon, Korea). Each DNA sequence of both strands was cross-checked using the universal primer sets SP6 and T7 to verify the sequences. Two or three partial nucleotide sequences were linked to confirm the full TK and gB gene sequences. Nucleotide sequence analysis was performed based on the full TK and gB genes of the FHV191071 and FHV191072 isolates, as well as other herpesviruses obtained from GenBank. The presumptive amino acid sequences of the TK proteins of the two isolates and the KANS-02 strain were compared with Clone Manager Basic ver. 9 (Sci-Ed Software, Denver, CO, USA). Two phylogenetic trees were constructed using MEGA ver. 7.0.20 (http://megasoftware.net). The bootstrap method with 1,000 replicates was used to assess the reliability of the phylogenetic trees.
Mouse inoculation test
A mouse experiment was performed at APQA, Korea using protocols approved by the experimental animal ethics committee (approval number: 2020-431). To evaluate the virulence of the FHV191071 and FHV191072 isolates, 18 four-week-old Balb/c mice (OrientBio, Seongnam, Korea) were divided into three groups. Two groups of six mice each were peritoneally inoculated with 108.0 TCID50/mL of the FHV191071 and FHV191072 isolates. The third group served as a control; mice in that group received no treatment. Mice were observed for clinical signs (e.g., itching, depression, and loss of appetite and body weight) for 10 days.
Go to : 
RESULTS
Isolation and biological characterization of the isolates
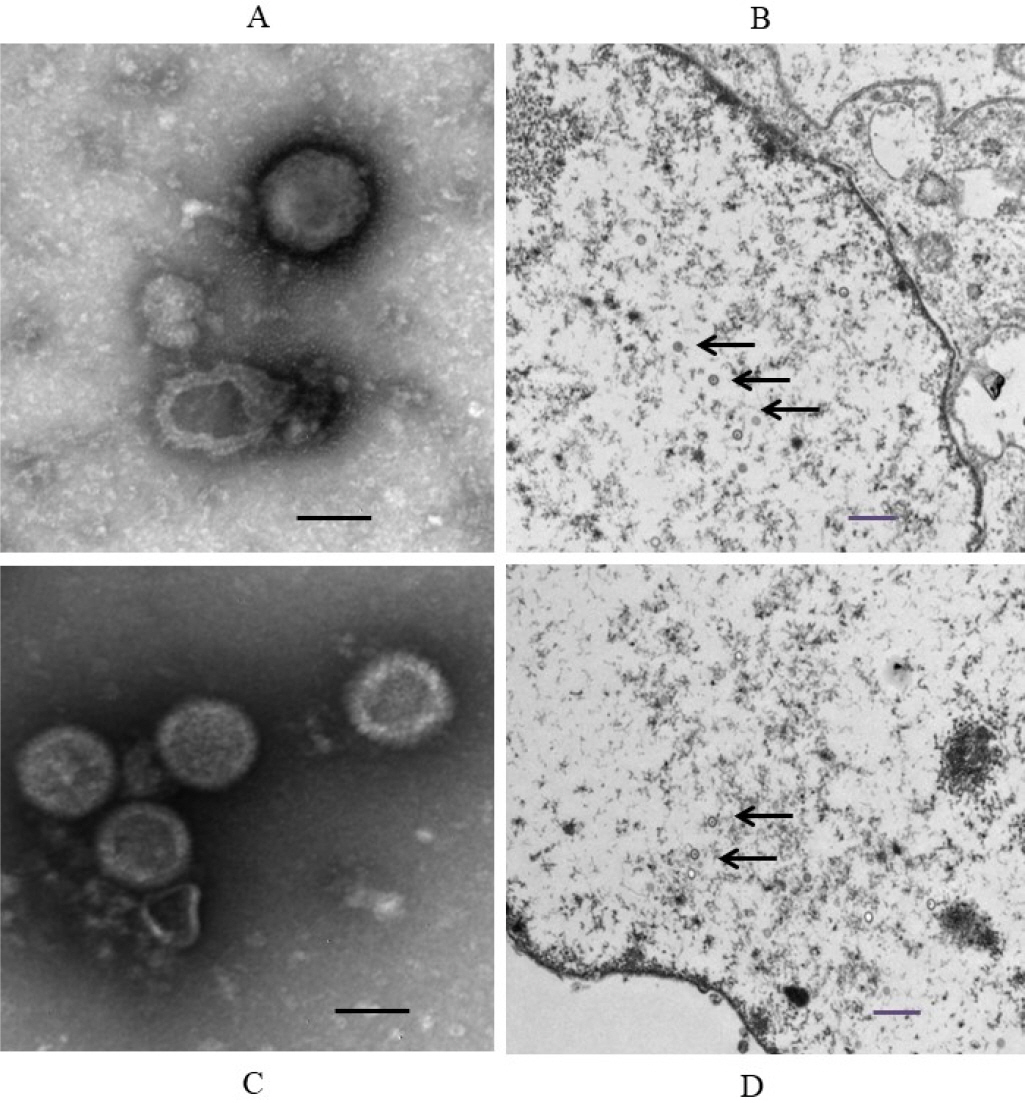
Of the CRFK cells inoculated with the 48 samples, only two samples from two cats exhibited specific CPE, including cell aggregation, detachment, and cell rounding (Fig. 1A, B). The samples with distinct CPE were named FHV191071 and FHV191072, respectively; CRFK cells infected with each isolate were fixed in cold acetone and reacted with mouse monoclonal antibodies against FHV-1. As shown in Fig. 1D and E, specific fluorescence was observed in CRFK cells infected with FHV191071 and FHV191072. Growth kinetics were examined to determine when the isolates exhibited the highest titer. As shown in Fig. 2A, the highest viral titers (108.3 and 108.5 TCID50/mL) were found when viruses were harvested 48 h after inoculation. Both isolates were inoculated into six cell types to assess cell tropism. Of the six cell types, both isolates exhibited the highest viral titers (range, 108.4 to 0 TCID50/mL) in CRFK cells and did not show any CPE in MDCK or Vero cells (Fig. 2B). HA assays were performed with three types of erythrocytes; neither isolate was able to agglutinate erythrocytes from cats, guinea pigs, or mice (data not shown). Based on the FHV191071 and FHV191072 growth kinetics, virus antigens for EM were prepared by sucrose purification. Virus particles of the FHV191071 and FHV191072 isolates were identified by EM from purified solutions by sucrose density gradient ultracentrifugation of infected cells. Viral particles with pleomorphic forms were 140–170 nm in diameter with crown-shaped projections, a morphological appearance typical of Herpesviridae (Fig. 3A, C). Viral particles were also observed in the nuclei of CRFK cells infected with both isolates (Fig. 3B, D).
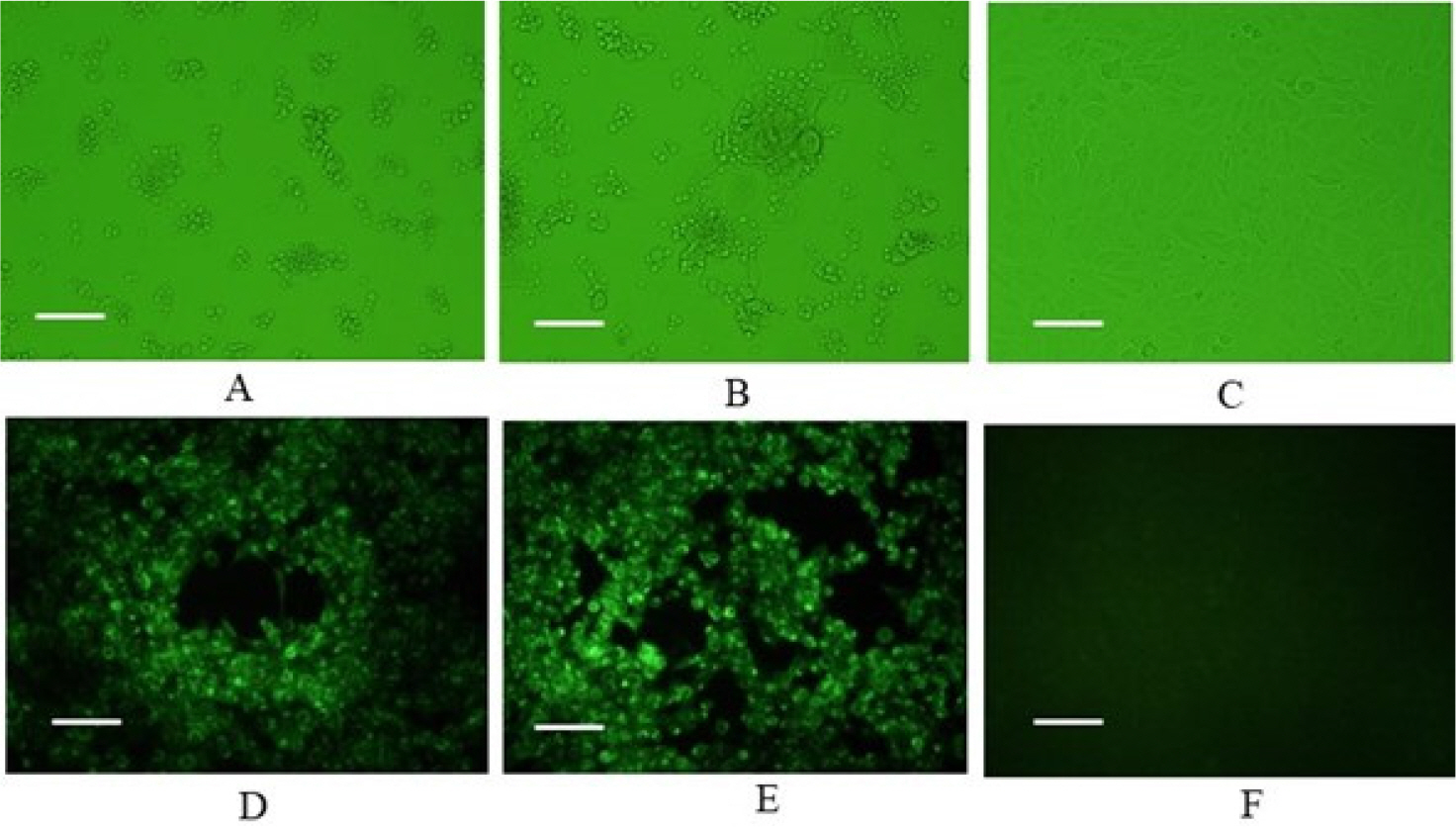 | Fig. 1Identification of feline herpesvirus type 1 (FHV-1) based on cytopathic effects (CPE) and fluorescence assays. CPE in CRFK cells infected with FHV191071 and FHV191072 isolates at 48 h post-inoculation (A, B); specific fluorescence observed in CRFK cells infected with FHV191071 and FHV191072 (D, E); and normal CRFK cells (C and F). Six pictures are magnified 200 times and scale bars indicate 100 μm. |
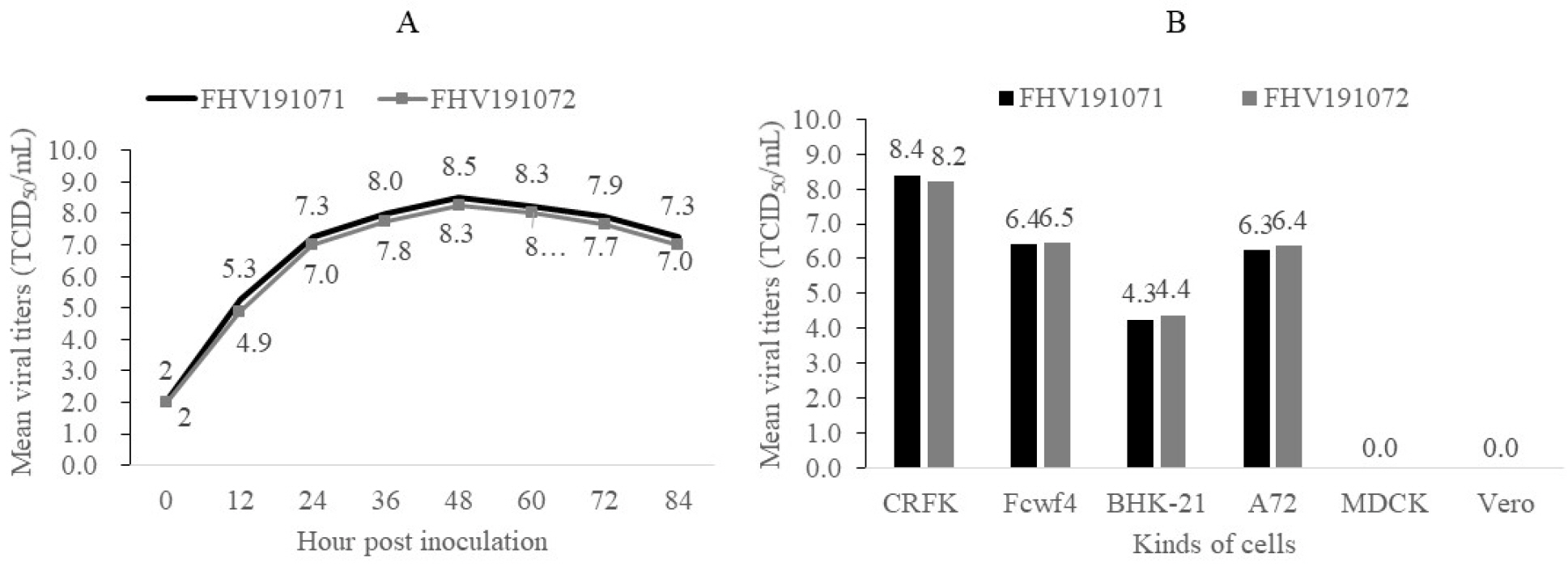 | Fig. 2Growth kinetics of FHV191071 and FHV191072 isolates, according to harvest time in CRFK cells and viral titers of two FHV isolates propagated in six cell types to examine cell tropism. The isolates showed maximum titers of 108.5 and 108.3 TCID50/mL in CRFK cells at 48 h post-inoculation (A). Mean viral titers of the two isolates ranged from 104.3 to 108.4 TCID50/mL in four cell types, while neither isolate grew in MDCK or Vero cells (B). |
Molecular characterization of FHV isolates
PCR, cloning, and sequencing were used to determine the genetic features of the two isolates. As shown in Fig. 4, six PCR products from the FHV191071 isolate were detected at sizes of 519, 620, 670, 889, 1109, and 1194 bp on 2.0% agarose gels, confirming that the isolates were FHV-1 based on the expected size of the PCR products. The entire TK and gB gene sequences of FHV191071 and FHV191072 isolates comprised 1,032 and 2,847 nucleotides encoding 344 and 949 amino acids, respectively. The TK and gB genes were compared with those of 16 herpesviruses in GenBank to determine their genetic relationships with other herpesviruses.
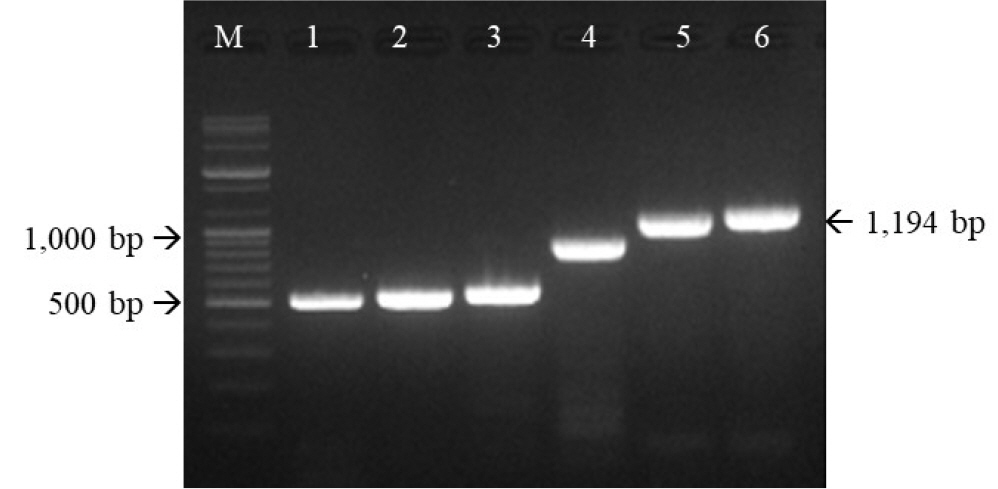 | Fig. 4Six primer sets targeting the thymidine kinase (TK) and glycoprotein B (gB) genes of the FHV191071 isolate were used for polymerase chain reaction (PCR). PCR products of expected sizes confirmed that the FHV191071 isolate was feline herpesvirus type 1. M, 1 kb ladder; PCR products of the TK (lanes 1-3) and gB (lanes 4-6) genes of the FHV191071 isolate. |
The TK and gB genes of the FHV191071 and FHV191072 isolates had identical nucleotide sequences and shared 99.9% to 100% nucleotide homology with those of other feline herpesviruses (e.g., KANS-02, KANA-04, and 729-83 strains reported in the USA and Australia) (11). Phylogenetic trees were constructed based on the two genes in 16 herpesviruses to examine the genetic relationships of the two isolates. As shown in Fig. 5, the herpesviruses were subdivided according to host animal species (i.e., swine, bovine, equine, feline, and canine); both isolates belonged to the feline group. The full amino acid sequences of the TK genes of FHV191071 and FHV1901072 isolates and the KANS-02 strain were aligned to examine genetic features of the two isolates. As shown in Fig. 6, one potential N-linked glycosylation site (N-X-S/T) was identified in the TK protein; one amino acid (isoleucine) was replaced by threonine at position 341 in the TK protein of both isolates.
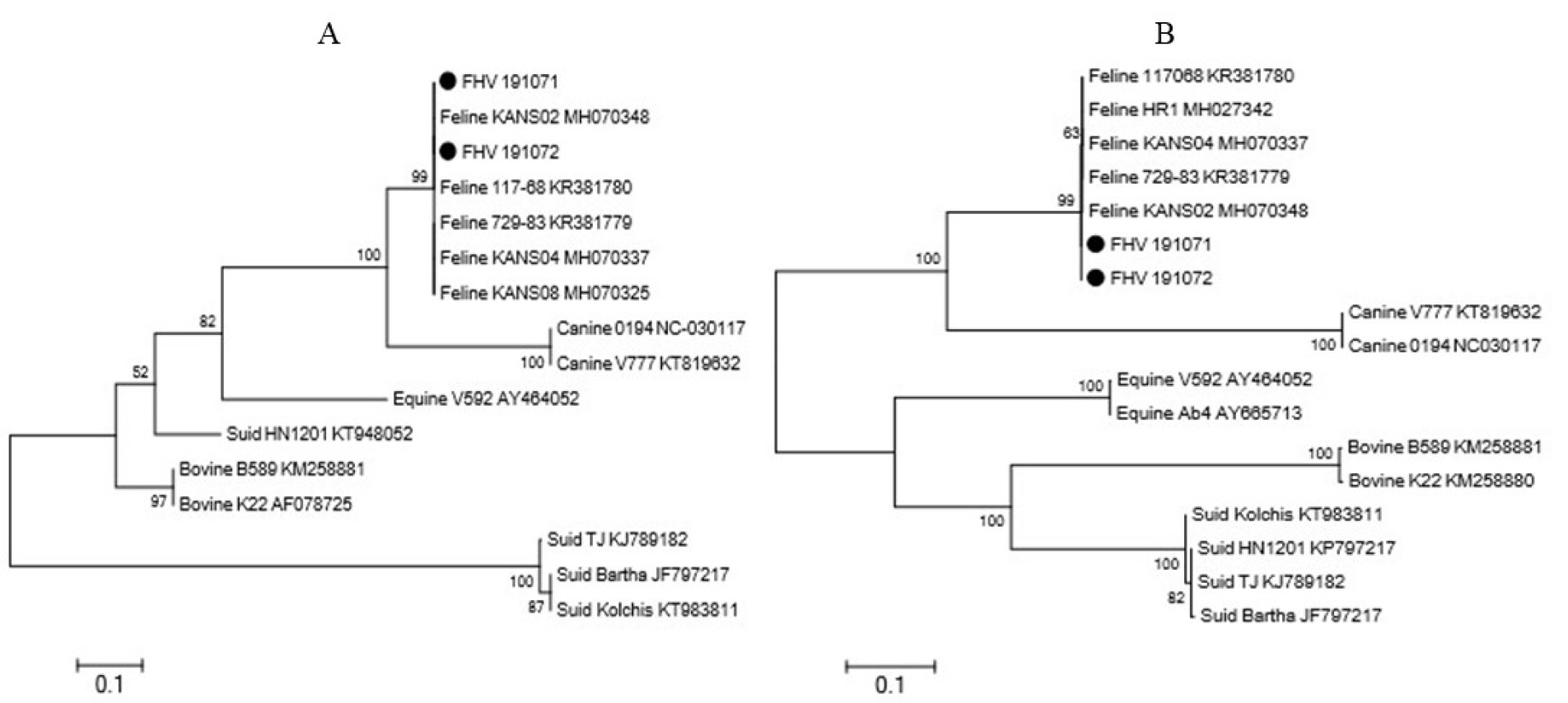 | Fig. 5Phylogenetic trees based on the thymidine kinase (TK) (A) and glycoprotein B (gB) genes of animal herpesviruses. In both phylogenetic trees, the FHV191071 and FHV191072 isolates belong to the feline herpesvirus type 1 group. |
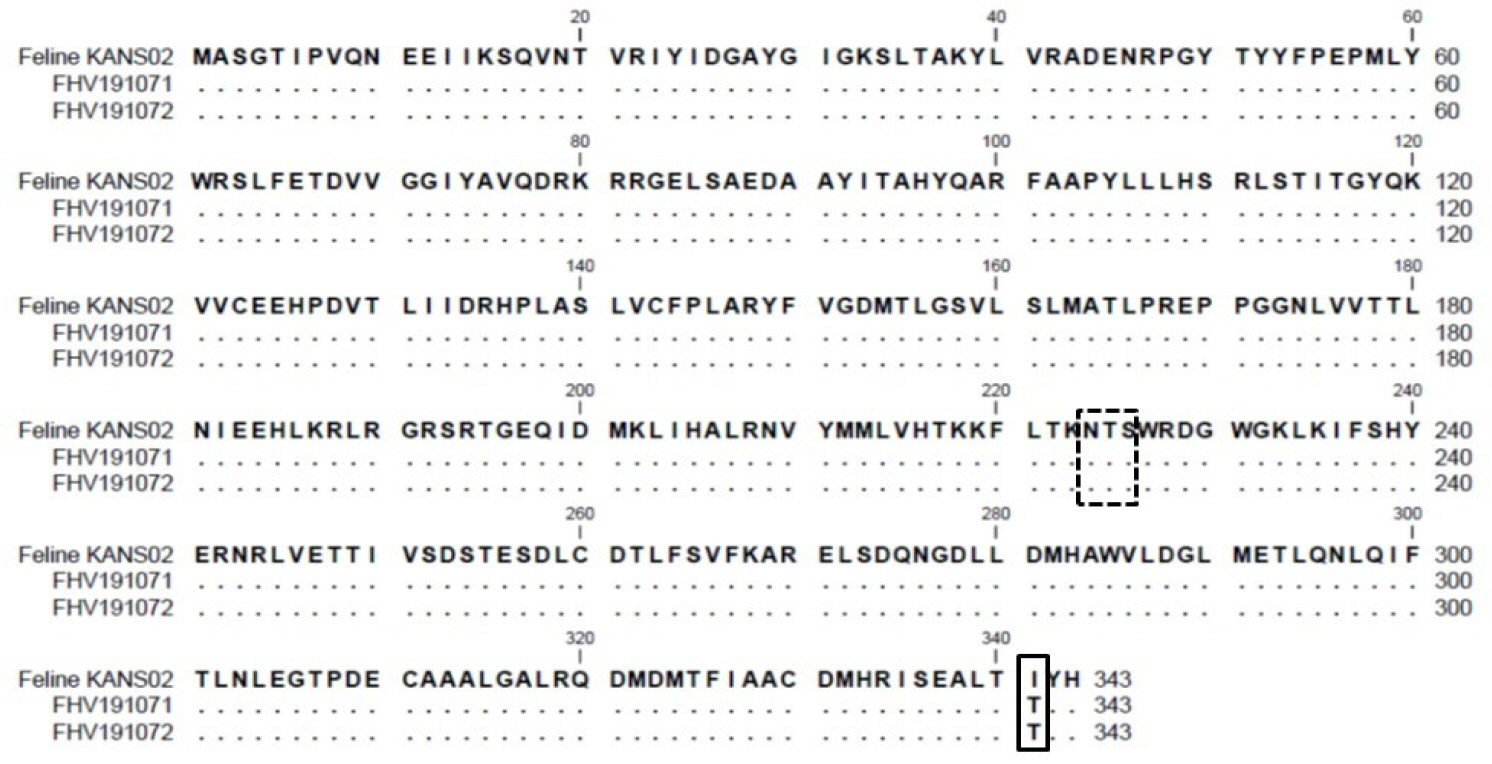 | Fig. 6Alignment of amino acid sequences of TK proteins of the FHV191071 and FHV191072 isolates and the KANS-02 strain. The amino acid threonine at position 341 in the TK protein of the Korean FHV isolates was replaced by isoleucine (solid box). A potential N-linked glycosylation site (N-X-S/T) is indicated by the box with a dashed line. |
Virulence of two isolates in mice
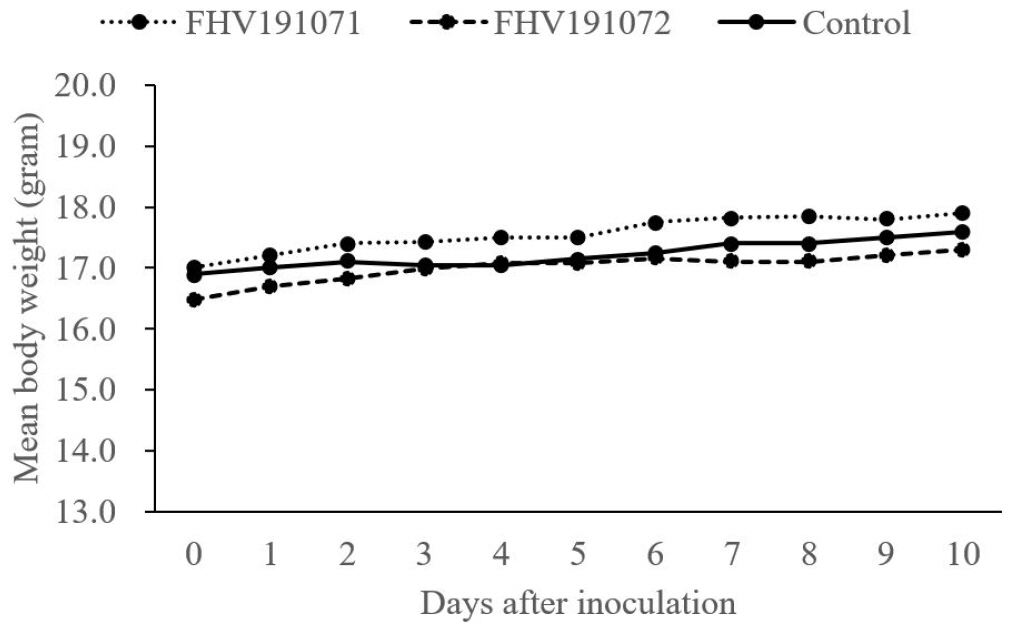
Six 4-week-old Balb/c mice were inoculated with FHV191071 and FHV191072 isolates at viral titers of 108.0 TCID50/mL to examine the virulence characteristics of the two isolates. None of the isolate-treated mice showed clinical signs or significant weight change during the observation period (Fig. 7).
Go to : 
DISCUSSION
The global distribution of FHV-1 has led to reports of FHV-1 companion cats and wild cats, including tigers, cheetahs, and lions (6, 7, 8, 15). Kittens infected with FHV-1 may die or exhibit severe clinical symptoms if they do not have maternal antibodies or are immunized with a vaccine that contains FHV-1 antigen (1, 5). Compared with other cat diseases, it is easy to diagnose FHV-1 in cats. Nevertheless, there have been few reports regarding the prevalence of FHV-1 in Korean cats or isolation of FHV-1 from these cats (16). From 12 dead cats sent to the APQA for diagnosis in 2019, we prepared 48 samples (kidney, brain, spleen, lymph nodes, lung, heart, liver, and intestine); we isolated two causative agents from kidney and brain homogenates of two cats, and cultured them in CRFK cells. These isolates were designated FHV191071 and FHV191072, respectively. We confirmed that both isolates were FHV-1 based on specific cell CPE, immunofluorescent assay, EM, PCR, and nucleotide sequence analysis. We attempted to identify histological lesions in the diagnosed cats by hematoxylin and eosin staining; however, no typical FHV-1 lesions were present. This may be early infection or latent infection. The most common laboratory diagnostic methods for FHV-1 are virus isolation and PCR using oronasal and conjunctival swabs, which yield DNA extracts (15, 17). Therefore, it is preferable to apply diagnostic methods including virus isolation and PCR to oronasal discharges of animals with suspected infection.
In this study, we found that CRFK cells produced the highest titers of the FHV-1 isolates among six cell lines, while MDCK and Vero cells showed no growth. Viral titers of the isolates reached 108.5 and 108.3 TCID50/mL 48 after inoculation, similar to wild-type FHV-1, C-27 strain (ATCC, Manassas, VA, USA) (2); this finding implied that the FHV191071 and FHV191072 isolates constituted wild-type virus circulating in stray cats. EM analysis of purified virus and infected CRFK cells revealed typical herpesvirus morphology. FHV191071 and FHV191072 particles were similar in size and shape to those of herpesviruses isolated from animals (15, 18). PCR detection methods (e.g., real-time and priming isothermal amplification assays) have previously been used to detect FHV-1 (4, 12). PCR amplification of FHV-1 based on the TK and gB genes yielded bands of the expected size, which confirmed the identity of FHV-1.
Phylogenetic analyses of FHV-1 TK and gB genes have been used to study evolutionary relationships and identify genetic variation among herpesviruses (1). When the TK genes of 16 herpesviruses were compared, the TK genes of both isolates were very similar to those of FHV-1. Sequence analyses of the gB genes of 15 herpesviruses, including both isolates, revealed that both isolates were in the FHV-1 group and exhibited comparable similarity to gB in other FHV-1 strains (e.g., KANS-02 and HR1). The reported genetic distance among FHV-1 stains is very low (0.093%) and the TK and gB genes are highly conserved within Varicellovirus (1, 15); the results regarding Korean FHV-1 isolates were consistent with previous results.
It has been well known that TK gene of Alphaherpesviruses is one of virulence factors and TK proteins are highly divergent. Alignment of the full amino acid sequences of TK proteins from both isolates with the same region from the KANS-02 strain revealed only one amino acid difference among the three FHV-1s, at position 341. Nunberg et al. reported that the TK gene was nonessential for herpesvirus growth and that loss of TK activity was involved in the attenuation of herpesvirus (9). The loss of function of the TK or gE gene provided the concept of a differential herpesvirus vaccine. Thus, FHV-1 with missing or mutated parts of the TK gene can be used as a strain marker. The isolation of FHV-1 from naturally infected cats is the first step in the development of a new FHV-1 vaccine, using FHV-1 isolated from Korean cats. A future study should sequence the entire genome of the Korean FHV-1 isolates or passaged virus.
In a mouse model, the Korean FHV-1 isolates did not cause any clinical signs in 4-week-old mice inoculated peritoneally, implying that factors associated with the alpha/beta interferon system are responsible for the replication of FHV-1 in mice. Bovine herpesvirus 1 and 5 can cause neurovirulence in interferon receptor-deficient mice (19). We found no clinical evidence that the FHV191071 and FHV191072 isolates were virulent in 4-week-old mice. Therefore, further studies regarding the virulence and immunogenicity of the Korean isolates are required in target animals, such as cats.
In conclusion, we isolated two viruses from naturally infected cats using CRFK cells and confirmed that they were FHV-1 closely related to the KANS-02 strain, which has been reported in the USA. CRFK cells produced the highest titers of wild FHV-1. The FHV191071 and FHV191072 isolates may provide raw material for use in developing a new FHV-1 vaccine and diagnostic reagents.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download