INTRODUCTION
METHODS
Human subjects
Table 1.
| Characteristic |
Microarray screening |
qRT-PCR validation |
||
|---|---|---|---|---|
| Control (n=3) | T1DM (n=3) | Control (n=12) | T1DM (n=12) | |
| Age, yr | 16 (12–18) | 15 (12–17) | 12.6 (6–18) | 12.8 (5–17) |
| Gender, female/male | 0/3 | 2/1 | 6/6 | 7/5 |
| Disease duration, mo | - | 3.7±2.1 | - | 3.0±1.9 |
| HbA1c, % | 5.6±0.1 | 9.7±0.8 | 5.4±0.4 | 9.9±1.2 |
| Subjects positive for ≥1 islet Aba | 0 | 3 | 0 | 12 |
| Serum C-peptide during OGTT, ng/mLb | ||||
| 0 min | - | 0.16±0.15 | - | 0.19±0.18 |
| 60 min | - | 0.21±0.18 | - | 0.39±0.37 |
| 120 min | - | 0.3±0.41 | - | 0.54±0.38 |
Values are presented as median (range) or mean±standard deviation.
qRT-PCR, quantitative real-time polymerase chain reaction; T1DM, type 1 diabetes mellitus; HbA1c, glycosylated hemoglobin; Ab, autoantibody; OGTT, oral glucose tolerance test.
RNA extraction, sample preparation, and microarray hybridization
Microarray data analysis
Prediction of miRNA response elements
Quantitative real-time polymerase chain reaction validation
Construction of the circRNA-miRNA-mRNA network
GO and KEGG analyses
Statistical analysis
RESULTS
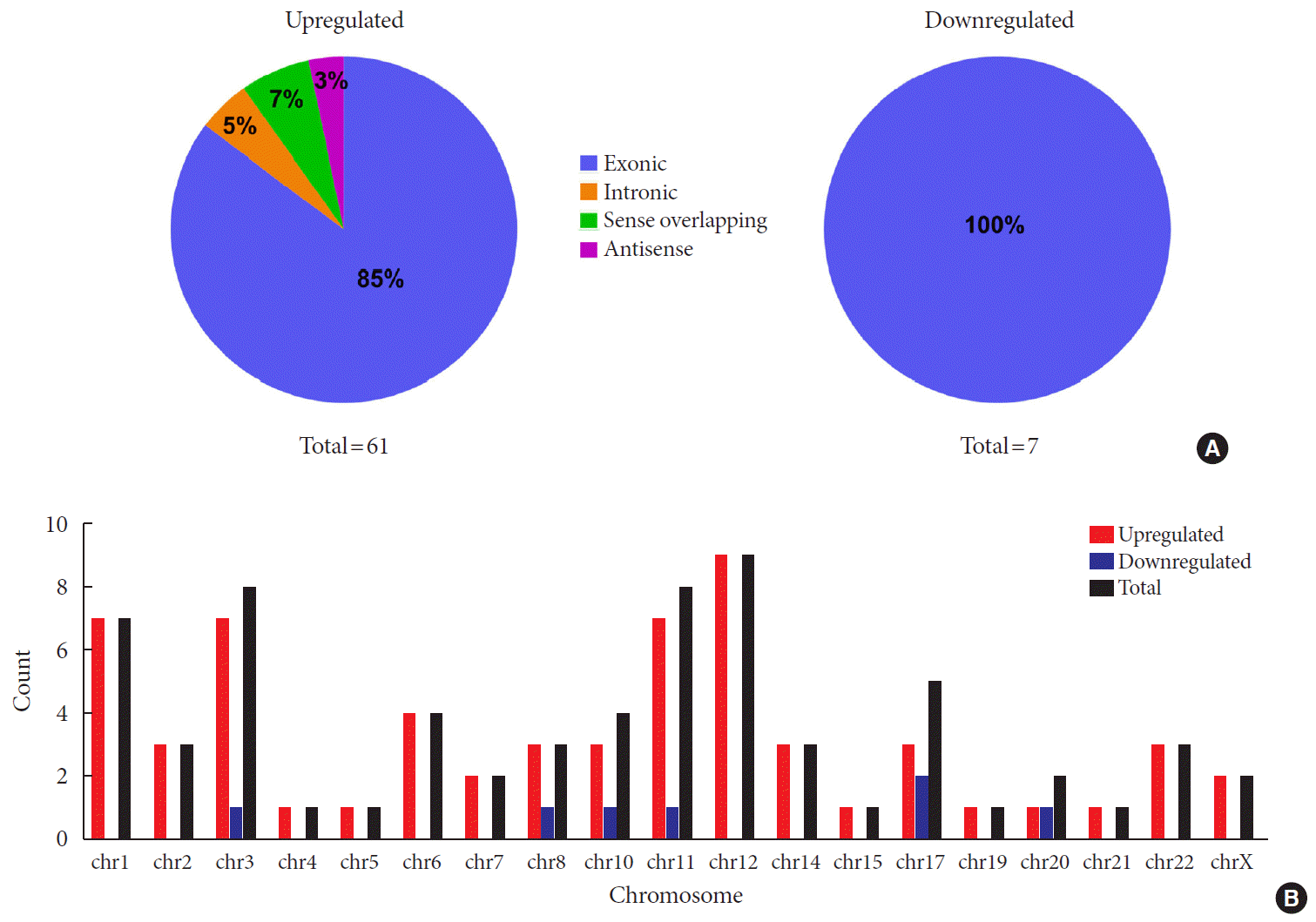
Identification of differentially expressed plasma circRNAs by new onset T1DM
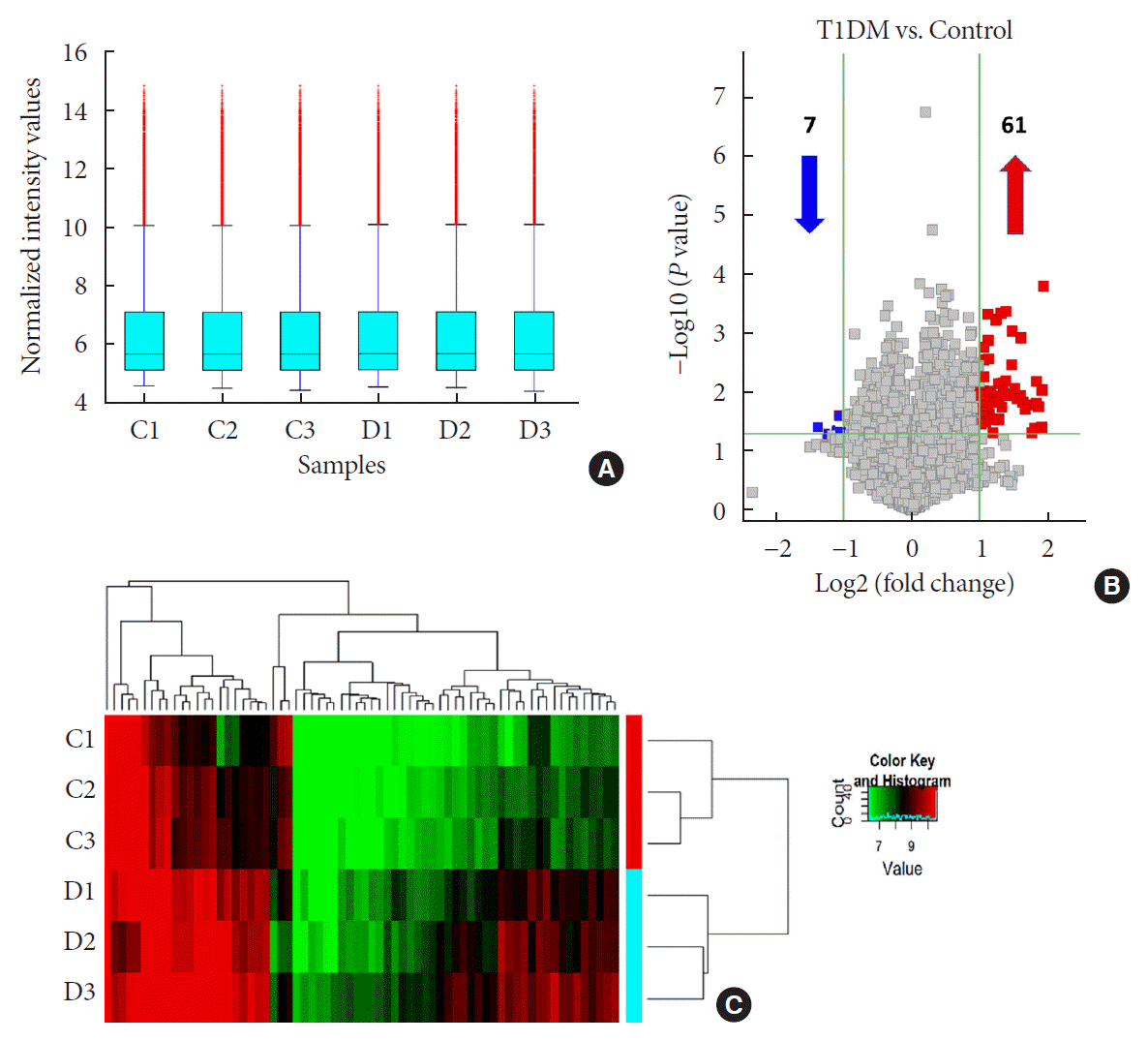
Fig. 1
Overview of differentially expressed plasma circular RNAs (circRNAs) identified in patients with newly diagnosed type 1 diabetes mellitus (T1DM) by microarray. (A) The box plot shows intensity distribution of expressed circRNA across all the samples after normalization. The central line within each box represents the median of the data, whereas the error bars represent the upper and lower quartiles. (B) Volcano plots show differentially expressed plasma circRNAs with fold-change greater than 2 and P value less than 0.05 between control and T1DM subjects. The upwards and downwards arrows indicate up- and down-regulated circRNA clusters, respectively. (C) Hierarchical cluster analysis (heat map) for visualizing differentially expressed circRNAs, wherein red and green colors denote high and low expression levels, respectively. C1–C3, healthy controls; D1–D3, T1DM patients.
Validation of differentially expressed circRNAs by qRT-PCR
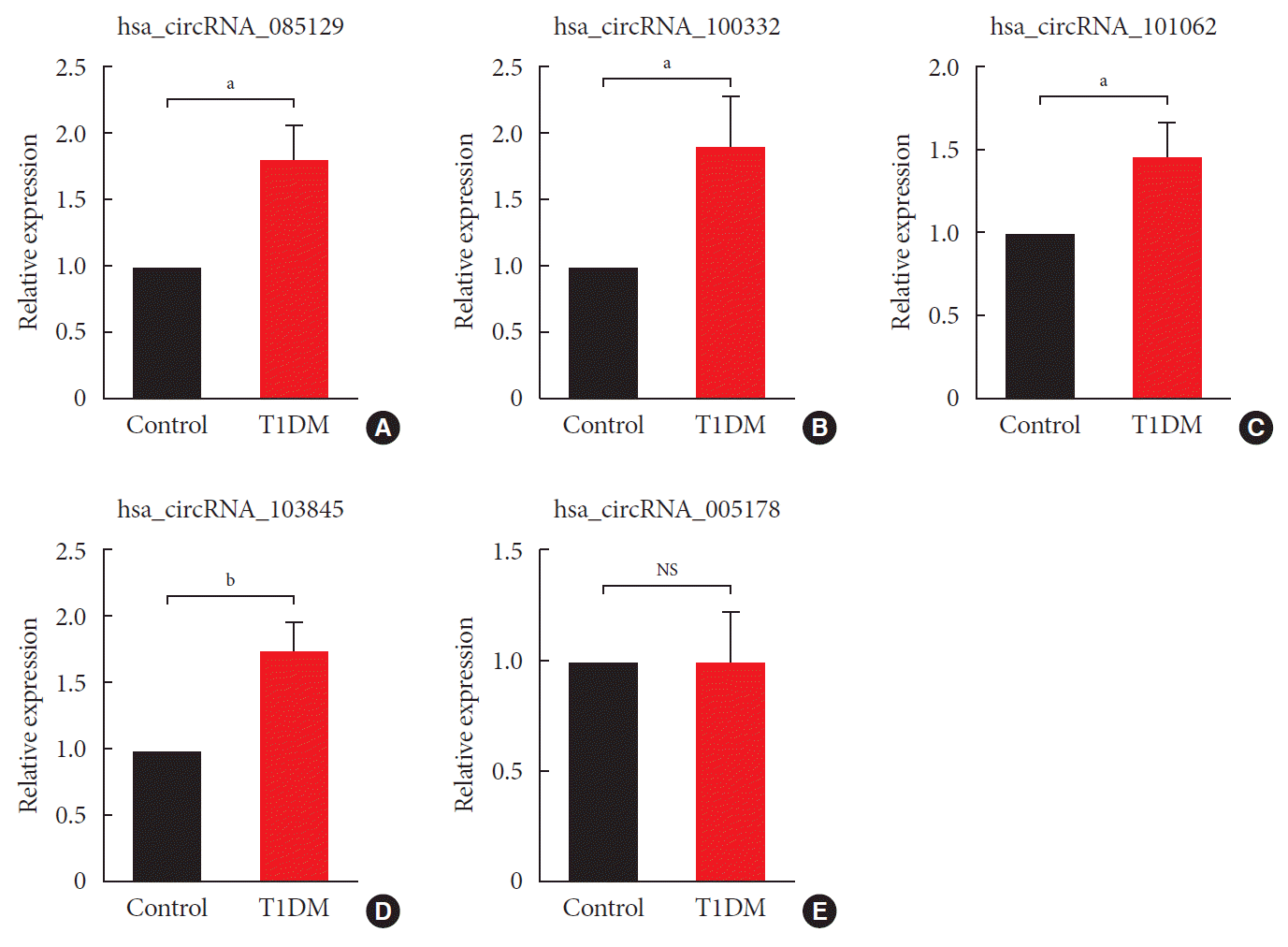
Fig. 3
Verification of microarray data by quantitative real-time polymerase chain reaction. Using independent human samples, our results confirmed that hsa_circular RNA (circRNA)_085129 (A), hsa_circRNA_100332 (B), hsa_circRNA_101062 (C) and hsa_circRNA_103845 (D) were all upregulated in type 1 diabetes mellitus (T1DM). However, expression of hsa_circRNA_005178 (E) remained unchanged. circRNA, circular RNA. Data are expressed as fold-change over healthy controls and represented as the mean±standard error (n=12). NS, not significant. aStatistically significant difference (P<0.05), bStatistically significant difference (P<0.01).
Prediction of miRNA response elements in differentially expressed circRNAs
Construction of circRNA-miRNA-mRNA network
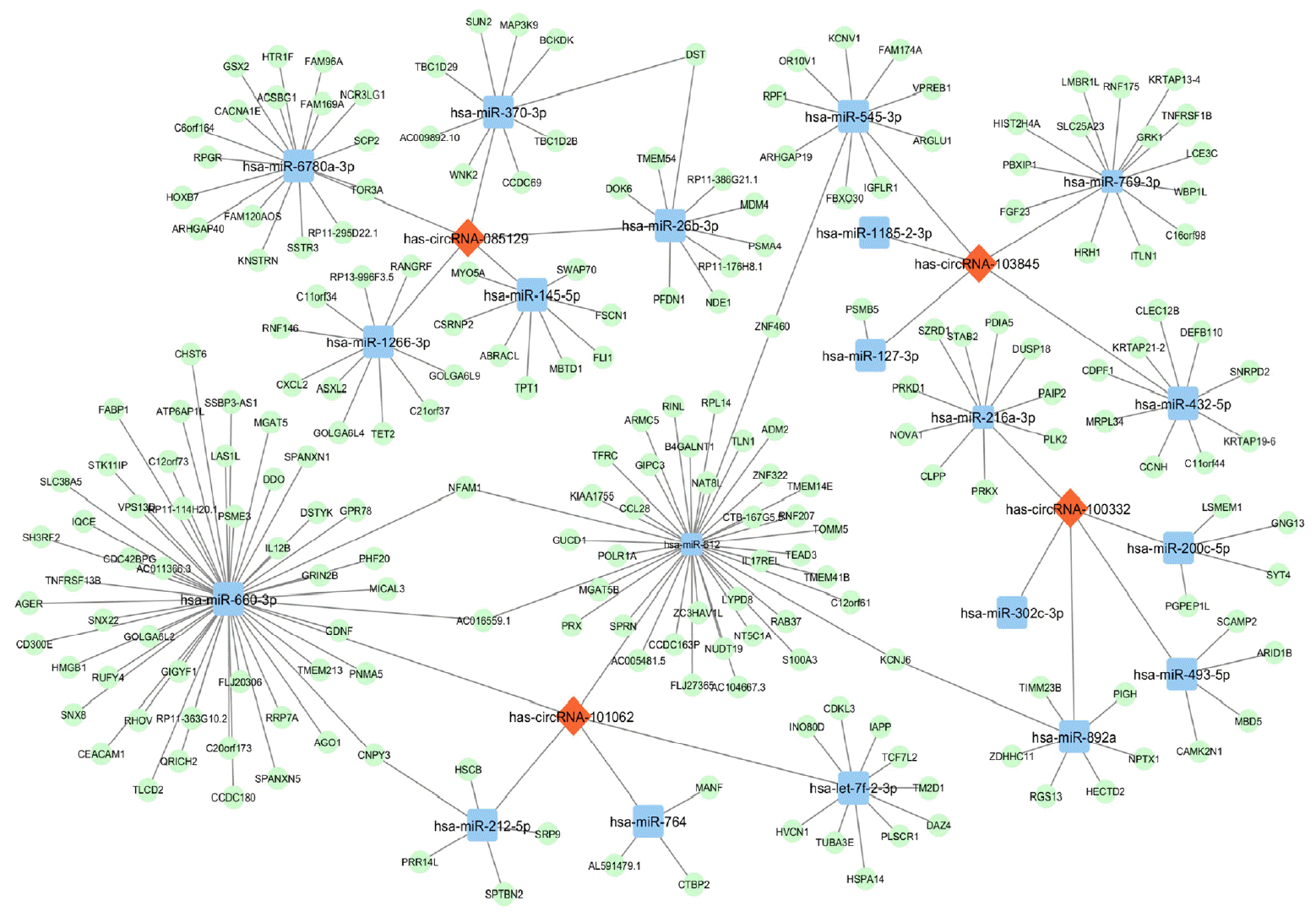
Fig. 4
The circular RNA (circRNA)-microRNA (miRNA)-messenger RNA (mRNA) network for the validated four circRNAs. Each circRNA interacts with its five miRNA response elements (MREs) and representative downstream mRNAs. For the convenience of visualization, only mRNAs with cumulative weighted context++ score no more than −0.7 were included. Specifically, hsa-miR-660-3p had the largest number of downstream target genes and hsa-miR-5189-5p exhibited the most interactions with other circRNA clusters. Red diamonds, circRNAs. Blue squares, miRNAs. Green ovals, mRNAs.
GO and KEGG pathway analyses
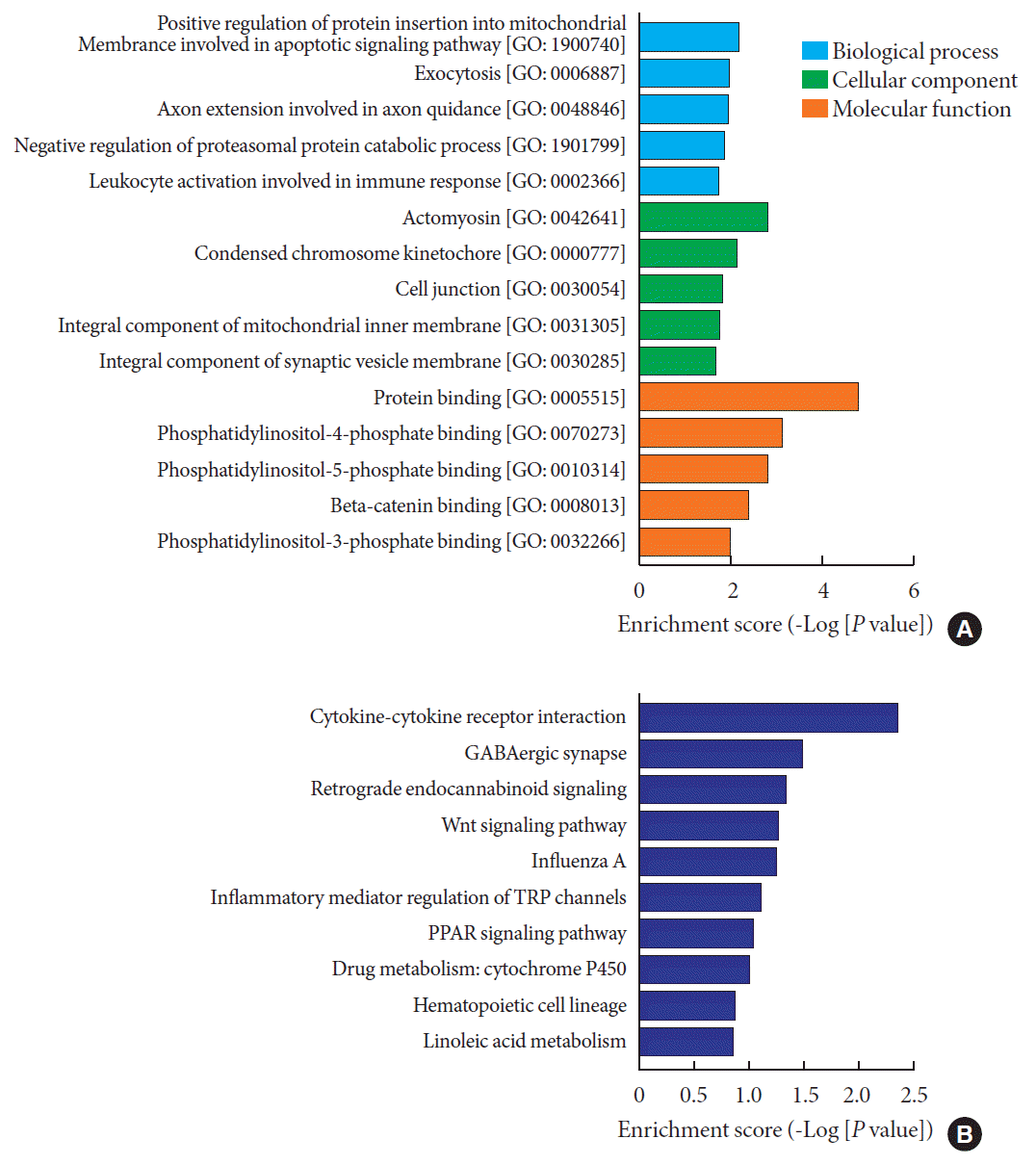
Fig. 5
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using predicted target messenger RNAs (mRNAs) of validated four circular RNA (circRNA). (A) GO enrichment analysis comprises three categories: biological process (in blue), cellular component (in green) and molecular function (in orange). Top five terms of each category are displayed. (B) KEGG pathway analysis shows top 10 terms that may be involved in the regulatory network mediated by differentially expressed circRNAs in type 1 diabetes mellitus.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download