2. Joseph KR, Edirimanne S, Eslick GD. 2018; Multifocality as a prognostic factor in thyroid cancer: A meta-analysis. Int J Surg. 50:121–5. DOI:
10.1016/j.ijsu.2017.12.035. PMID:
29337178.

3. Kim HJ, Sohn SY, Jang HW, Kim SW, Chung JH. 2013; Multifocality, but not bilaterality, is a predictor of disease recurrence/persistence of papillary thyroid carcinoma. World J Surg. 37(2):376–84. DOI:
10.1007/s00268-012-1835-2. PMID:
23135422.

4. Park SY, Park YJ, Lee YJ, Lee HS, Choi SH, Choe G, et al. 2006; Analysis of differential BRAF(V600E) mutational status in multifocal papillary thyroid carcinoma: evidence of independent clonal origin in distinct tumor foci. Cancer. 107(8):1831–8. DOI:
10.1002/cncr.22218. PMID:
16983703.
5. Iacobone M, Jansson S, Barczynski M, Goretzki P. 2014; Multifocal papillary thyroid carcinoma--a consensus report of the European Society of Endocrine Surgeons (ESES). Langenbecks Arch Surg. 399(2):141–54. DOI:
10.1007/s00423-013-1145-7. PMID:
24263684.

6. Wang W, Su X, He K, Wang Y, Wang H, Wang H, et al. 2016; Comparison of the clinicopathologic features and prognosis of bilateral versus unilateral multifocal papillary thyroid cancer: An updated study with more than 2000 consecutive patients. Cancer. 122(2):198–206. DOI:
10.1002/cncr.29689. PMID:
26506214.

7. Amin A, Younis G, Sayed K, Saeed Z. 2015; Cervical lymph node metastasis in differentiated thyroid carcinoma: does it have an impact on disease-related morbid events? Nucl Med Commun. 36(2):120–4. DOI:
10.1097/MNM.0000000000000234. PMID:
25356622.
8. Blanchard C, Brient C, Volteau C, Sebag F, Roy M, Drui D, et al. 2013; Factors predictive of lymph node metastasis in the follicular variant of papillary thyroid carcinoma. Br J Surg. 100(10):1312–7. DOI:
10.1002/bjs.9210. PMID:
23939843.

9. Pinyi Z, Bin Z, Jianlong B, Yao L, Weifeng Z. 2014; Risk factors and clinical indication of metastasis to lymph nodes posterior to right recurrent laryngeal nerve in papillary thyroid carcinoma: a single-center study in China. Head Neck. 36(9):1335–42. DOI:
10.1002/hed.23451. PMID:
23955992.

10. Singhal S, Sippel RS, Chen H, Schneider DF. 2014; Distinguishing classical papillary thyroid microcancers from follicular-variant microcancers. J Surg Res. 190(1):151–6. DOI:
10.1016/j.jss.2014.03.032. PMID:
24735716. PMCID:
PMC4053495.

11. Konturek A, Barczynski M, Nowak W, Richter P. 2012; Prognostic factors in differentiated thyroid cancer--a 20-year surgical outcome study. Langenbecks Arch Surg. 397(5):809–15. DOI:
10.1007/s00423-011-0899-z. PMID:
22350610. PMCID:
PMC3349847.

12. Ahn D, Sohn JH, Kim JH, Shin CM, Jeon JH, Park JY. 2013; Preoperative subclinical hypothyroidism in patients with papillary thyroid carcinoma. Am J Otolaryngol. 34(4):312–9. DOI:
10.1016/j.amjoto.2012.12.013. PMID:
23357595.

15. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998; Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 95(25):14863–8. DOI:
10.1073/pnas.95.25.14863. PMID:
9843981. PMCID:
PMC24541.

16. Kim SJ, Myong JP, Jee HG, Chai YJ, Choi JY, Min HS, et al. 2016; Combined effect of Hashimoto's thyroiditis and BRAF(V600E) mutation status on aggressiveness in papillary thyroid cancer. Head Neck. 38(1):95–101. DOI:
10.1002/hed.23854. PMID:
25213729.
17. Bircan HY, Koc B, Akarsu C, Demiralay E, Demirag A, Adas M, et al. 2014; Is Hashimoto's thyroiditis a prognostic factor for thyroid papillary microcarcinoma? Eur Rev Med Pharmacol Sci. 18(13):1910–5. PMID:
25010622.
18. Lee YK, Park KH, Park SH, Kim KJ, Shin DY, Nam KH, et al. 2018; Association between diffuse lymphocytic infiltration and papillary thyroid cancer aggressiveness according to the presence of thyroid peroxidase antibody and BRAF(V600E) mutation. Head Neck. 40(10):2271–9. DOI:
10.1002/hed.25327. PMID:
29935011.

19. Qu HJ, Qu XY, Hu Z, Lin Y, Wang JR, Zheng CF, et al. 2018; The synergic effect of BRAF(V600E) mutation and multifocality on central lymph node metastasis in unilateral papillary thyroid carcinoma. Endocr J. 65(1):113–20. DOI:
10.1507/endocrj.EJ17-0110. PMID:
29070763.
20. Kimbrell HZ, Sholl AB, Ratnayaka S, Japa S, Lacey M, Carpio G, et al. 2015; BRAF testing in multifocal papillary thyroid carcinoma. Biomed Res Int. 2015:486391. DOI:
10.1155/2015/486391. PMID:
26448939. PMCID:
PMC4584030.
21. Giordano D, Gradoni P, Oretti G, Molina E, Ferri T. 2010; Treatment and prognostic factors of papillary thyroid microcarcinoma. Clin Otolaryngol. 35(2):118–24. DOI:
10.1111/j.1749-4486.2010.02085.x. PMID:
20500581.

22. Hay ID, Hutchinson ME, Gonzalez-Losada T, McIver B, Reinalda ME, Grant CS, et al. 2008; Papillary thyroid microcarcinoma: a study of 900 cases observed in a 60-year period. Surgery. 144(6):980–7. discussion 7–8. DOI:
10.1016/j.surg.2008.08.035. PMID:
19041007.

23. Lombardi CP, Bellantone R, De Crea C, Paladino NC, Fadda G, Salvatori M, et al. 2010; Papillary thyroid microcarcinoma: extrathyroidal extension, lymph node metastases, and risk factors for recurrence in a high prevalence of goiter area. World J Surg. 34(6):1214–21. DOI:
10.1007/s00268-009-0375-x. PMID:
20052467.

24. Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, et al. 2013; Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 309(14):1493–501. DOI:
10.1001/jama.2013.3190. PMID:
23571588. PMCID:
PMC3791140.

25. Henke LE, Pfeifer JD, Ma C, Perkins SM, DeWees T, El-Mofty S, et al. 2015; BRAF mutation is not predictive of long-term outcome in papillary thyroid carcinoma. Cancer Med. 4(6):791–9. DOI:
10.1002/cam4.417. PMID:
25712893. PMCID:
PMC4472201.
26. Gouveia C, Can NT, Bostrom A, Grenert JP, van Zante A, Orloff LA. 2013; Lack of association of BRAF mutation with negative prognostic indicators in papillary thyroid carcinoma: the University of California, San Francisco, experience. JAMA Otolaryngol Head Neck Surg. 139(11):1164–70. DOI:
10.1001/jamaoto.2013.4501. PMID:
24030686.
27. Kim HK, Lee I, Lee J, Chang HS, Soh EY, Park IS, et al. 2018; BRAF(wild) papillary thyroid carcinoma has two distinct mRNA expression patterns with different clinical behaviors. Head Neck. 40(8):1707–18. DOI:
10.1002/hed.25151. PMID:
29573027.
29. An JH, Song KH, Kim SK, Park KS, Yoo YB, Yang JH, et al. 2015; RAS mutations in indeterminate thyroid nodules are predictive of the follicular variant of papillary thyroid carcinoma. Clin Endocrinol (Oxf). 82(5):760–6. DOI:
10.1111/cen.12579. PMID:
25109485.

30. Oh KH, Jung KY, Baek SK, Woo JS, Cho JG, Kwon SY. 2017; Relation between RASSF1A methylation and BRAF mutation in thyroid tumor. Int J Thyroidol. 11(2):123–9. DOI:
10.11106/ijt.2018.11.2.123.

31. Mauer J, Denson JL, Bruning JC. 2015; Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 36(2):92–101. DOI:
10.1016/j.it.2014.12.008. PMID:
25616716.

32. Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, et al. 2010; Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 115(23):4742–9. DOI:
10.1182/blood-2009-10-249540. PMID:
20351312. PMCID:
PMC2890190.

33. Jovanovic L, Delahunt B, McIver B, Eberhardt NL, Grebe SK. 2008; Most multifocal papillary thyroid carcinomas acquire genetic and morphotype diversity through subclonal evolution following the intra-glandular spread of the initial neoplastic clone. J Pathol. 215(2):145–54. DOI:
10.1002/path.2342. PMID:
18393366.

34. Lin X, Finkelstein SD, Zhu B, Silverman JF. 2008; Molecular analysis of multifocal papillary thyroid carcinoma. J Mol Endocrinol. 41(4):195–203. DOI:
10.1677/JME-08-0063. PMID:
18628356.

35. McCarthy RP, Wang M, Jones TD, Strate RW, Cheng L. 2006; Molecular evidence for the same clonal origin of multifocal papillary thyroid carcinomas. Clin Cancer Res. 12(8):2414–8. DOI:
10.1158/1078-0432.CCR-05-2818. PMID:
16638846.

36. Lu Z, Sheng J, Zhang Y, Deng J, Li Y, Lu A, et al. 2016; Clonality analysis of multifocal papillary thyroid carcinoma by using genetic profiles. J Pathol. 239(1):72–83. DOI:
10.1002/path.4696. PMID:
27071483. PMCID:
PMC5706659.

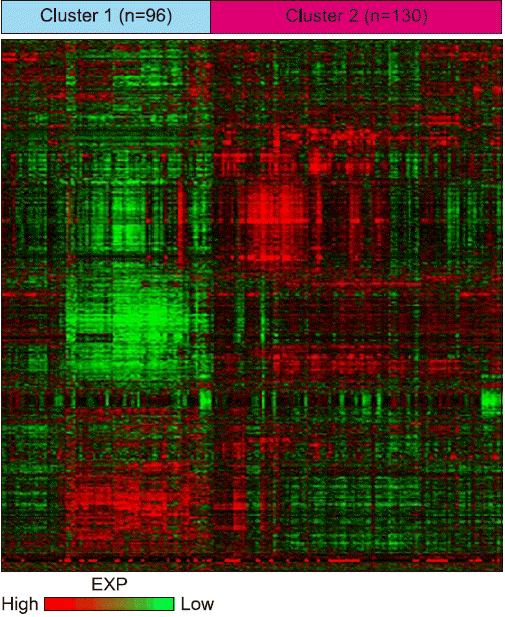
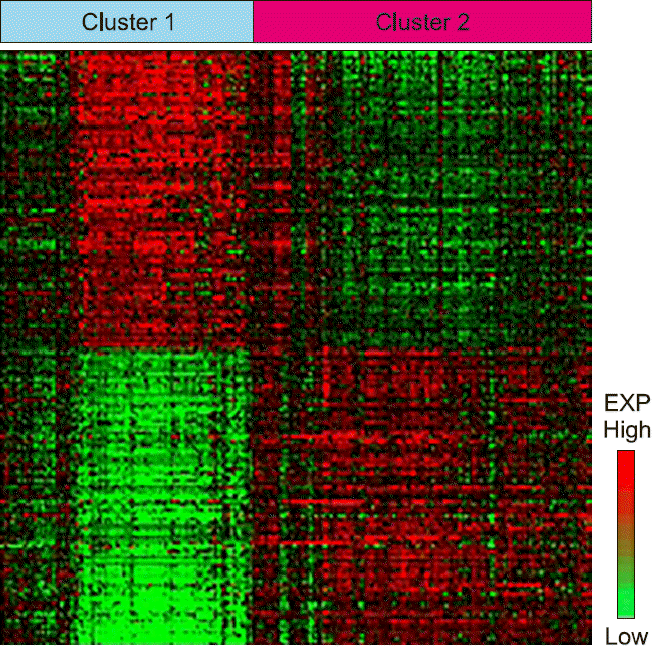




 PDF
PDF Citation
Citation Print
Print



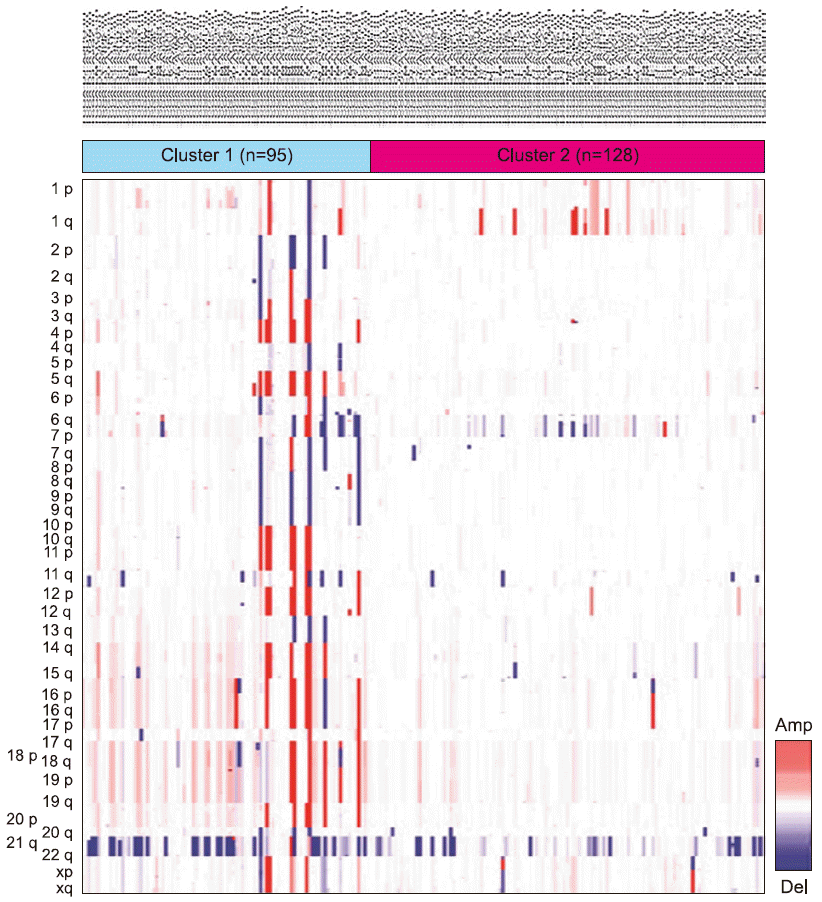
 XML Download
XML Download