High-quality biospecimens are necessary for biomedical research. Well-preserved frozen tissue is the favored biospecimen for biomedical research as it produces higher quality DNA, RNA, and proteins than formalin-fixed paraffin-embedded (FFPE) tissue.1 Biobanking aims to systemically collect, transport, store, manage, and utilize fresh tissues from surgical removal for further projects under the standard operation procedures (SOP).2 Therefore, biobanking plays an important role in clinical and translational research as well as future scientific research.
Preanalytical handling basically refers to all processes that occur until the analysis of a biospecimen.3 Preanalytical variables including ischemia time (IT) and storage period (SP) can have significant effects on the integrity of the biospecimens and the results of downstream analyses.3 In view of this, preanalytical variables must be precisely assessed and controlled in biobanking for optimal use of biospecimens in future.
Knowledge about acceptable cold ischemia time (CIT) (i.e., the time interval between surgical removal in the operating room and freezing of tissue at the pathology department)4 or SP is considered essential to improve the quality of biospecimens. Previous studies reported conflicting results regarding DNA quality for long CIT or SP.5–8 For colorectal cancer (CRC) tissues, a recent study reported minimal RNA change for up to 6 hours of CIT9 and tissues stored in the liquid nitrogen tank for 11 years can provide high-quality specimens for medical research.10 Over the past decade, there have been significant advances in the molecular characterization of various cancers that are driving treatment decisions. In CRC patients, KRAS is one of the most important biomarkers for identifying patients who will respond to anti-epidermal growth factor receptor (EGFR) therapies.11 In addition to KRAS, the number of molecular tests performed on CRC tissue specimen is constantly increasing. However, molecular diagnostics is confronted with its own specific challenges such as the amount of tissue specimens or archival status. Therefore, growing interest in molecular analysis in CRC can be facilitated by biobanked specimens.
Given the above considerations, the goal of this study was to define the impact of CIT and SP on DNA purity and integrity in biobanked CRC tissues. Furthermore, we assessed the concordance of KRAS mutation status between biobanked and FFPE tissues to evaluate the practical usability of DNA from a biobanked tissue for biomarker research. It is hypothesized that the results of this study will help to formulate appropriate guidelines for preanalytical processing of CRC specimens in biobanking.
MATERIALS AND METHODS
Biobanking of CRC tissue
Forty-five, fresh-frozen CRC tissues collected from 2013 to 2017 and stored at −80°C in Bio-Resource Bank of Dong-A University Medical Center (DAMC) were evaluated in order to define the influence of CIT and SP on DNA quality and biomarker research in biobanked human CRC tissues. Handling of resected CRC tissues from patients was carried out by trained and experienced staffs according to the SOP of Bio-Resource Bank of DAMC. In short, resection specimens were transported (at room temperature without any conservation fluid) from the operating room to the pathology department, immediately following removal of the specimen from the patient. In the pathology department, the specimen was handled at room temperature and within 60 minutes after resection specimens were snap-frozen as described below. When the 60 minutes time limit was exceeded, no tissue samples were taken. Blood and necrotic tissues on the surface of the specimens were washed off with pre-cooled phosphate-buffered saline (PBS). Subsequently, the tumor specimens were cut into small pieces (about 0.5 × 0.5 × 0.5 cm in dimension) under aseptic conditions. The frozen slides were observed by an experienced pathologist (M.S.Roh) and tumor specimens which comprised of ≥ 80% tumor nuclei and ≤ 20% necrosis, were considered as acceptable for future research. Tumors and normal tissues were repackaged in labeled cryovials and quickly frozen in −80°C freezer with specialized temperature monitoring system to direct real-time temperature.
Study design
CRC tissues were chosen according to three CITs (less than 30 minutes (CIT-1), 31 – 45 minutes (CIT-2), and 46–60 minutes (CIT-3)) and three SPs (less than 1 year (SP-1), 2–3 years (SP-2), and 4–5 years (SP-3)). Next, the specimens were assigned to nine groups (Table 1) and five specimens were allocated to each group. In all cases, molecular analysis for KRAS mutation status was initially performed using FFPE tissue block at the time of pathologic diagnosis. The specimens used were collected after obtaining informed consent from the donors and immediately anonymized. The study protocol was approved by the Institutional Review Board of DAMC of Korea (DAUHIRB-18-099).
Table 1
Mean 260/280 ratio for DNA purity analysis in nine groups assigned by combination of cold ischemia time and storage period
DNA preparation
Frozen tissues were prepared with optimal cutting temperature (OCT)-embedded tissue and cut into 5 μm pieces. The cut sections were washed with PBS and the OCT compound was removed. Cell lysis and protein removal were performed using a lysis buffer and proteinase K, respectively. DNA was extracted using the paramagnetic beads based Maxwell®16 Viral Total Nucleic Acid Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instruction.
DNA purity analysis by spectrophotometry
After eluting DNA in 50 μl of elution buffer, the concentration and purity of extracted DNA were assessed using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The machine was calibrated and cleaned according to the manufacturer’s instruction. Absorbance at 260 nm and 280 nm for 2 μl of each DNA specimen was measured. The corresponding elution buffer was used as blank. Due to DNA absorbance at a wavelength of 260 nm and protein absorbance at a wavelength of 280 nm, the 260/280 ratio was used as the purity indicator of the DNA specimens. Since an optimum value for 260/280 ratio for pure DNA is 1.8, the percentage of specimens for each group with a purity ratio between 1.6 and 2.0 (1.8 ± 0.2) was additionally measured. DNA purity was finally determined according to 260/280 ratio as follows; 260/280 ratio between 1.8 and 2.0 was accepted as pure for DNA, and less than 1.8 or 2.0 or more was judged as bad. A ratio lower than 1.8 was considered as indicative of the presence of protein, phenol, or other contaminants that absorb strongly at or near 280 nm. And ratio of more than 2.0 was assumed as indicative of RNA contamination.12
DNA integrity analysis by electrophoresis
To observe DNA integrity, 50 ng of each DNA specimen based on spectrophotometric measurement was analyzed by electrophoresis on a 2% agarose gel (Biosesang, Korea). Lambda DNA (Bioneer, Korea) and 1 Kb DNA ladder (Bioneer, Korea) were used as positive control and DNA molecular weight marker, respectively. After loading and running for 2 hours at a voltage of 50 V, the results were analyzed using a UV transilluminator. Well-preserved DNA appeared as a unique, well-defined high molecular weight band higher than 20 kb. DNA was determined to be degraded when smearing was observed.13
KRAS mutation status analysis by peptide nucleic acid (PNA) clamping
To evaluate the feasibility of KRAS molecular testing in biobanked specimens, KRAS exon 2 (codon12 and 13) mutation analysis by PNA-mediated real-time PCR clamping was performed on biobanked frozen tissue in all specimens. The amount of DNA used for PNA clamping was 40–80 ng/test (10 ng/reaction). A PNAClampTM KRAS Mutation Detection Kit (Panagene, Daejeon, Korea) was used to detect KRAS mutations by real-time PCR, according to the manufacturer’s instruction. Briefly, all reactions included template DNA, a primer and PNA probe set, and the SYBR Green PCR master mix. Real-time PCR was performed using a CFX96 PCR detection system (Bio-Rad, Philadelphia, PA, USA). The efficiency of PCR clamping was determined by measuring the threshold cycle (Ct) values. Higher ΔCt (ΔCt = Ctstandard – Ctspecimen) values meant that the mutant DNA was efficiently amplified. A cutoff value of 2.0 was used to indicate the presence of mutant DNA. KRAS mutation status of all 45 biobanked frozen tissues that was performed at this time was compared with the results of corresponding FFPE tissues that was initially performed at the time of pathologic diagnosis.
Go to : 
RESULTS
DNA purity of different CIT and SP
All DNA specimens had a 260/280 ratio ranging between 1.8 and 2.0 with the exception of a single specimen. The specimen with a ratio out of range belonged to group 5 (CIT-2/SP-2) and showed 1.76 of 260/280 ratio, suggesting the presence of protein or other contaminants that absorb strongly at or near 280 nm. DNA remained stable in CRC tissues kept at room temperature for up to 60 minutes (mean 260/280 purity ratio: 1.89 in CIT-1, 1.83 in CIT-2, and 1.90 in CIT-3) and long-term storage for up to 5 years (mean 260/280 ratio: 1.88 in SP-1, 1.87 in SP-2, and 1.87 in SP-3) (Table 1) (Fig. 1).
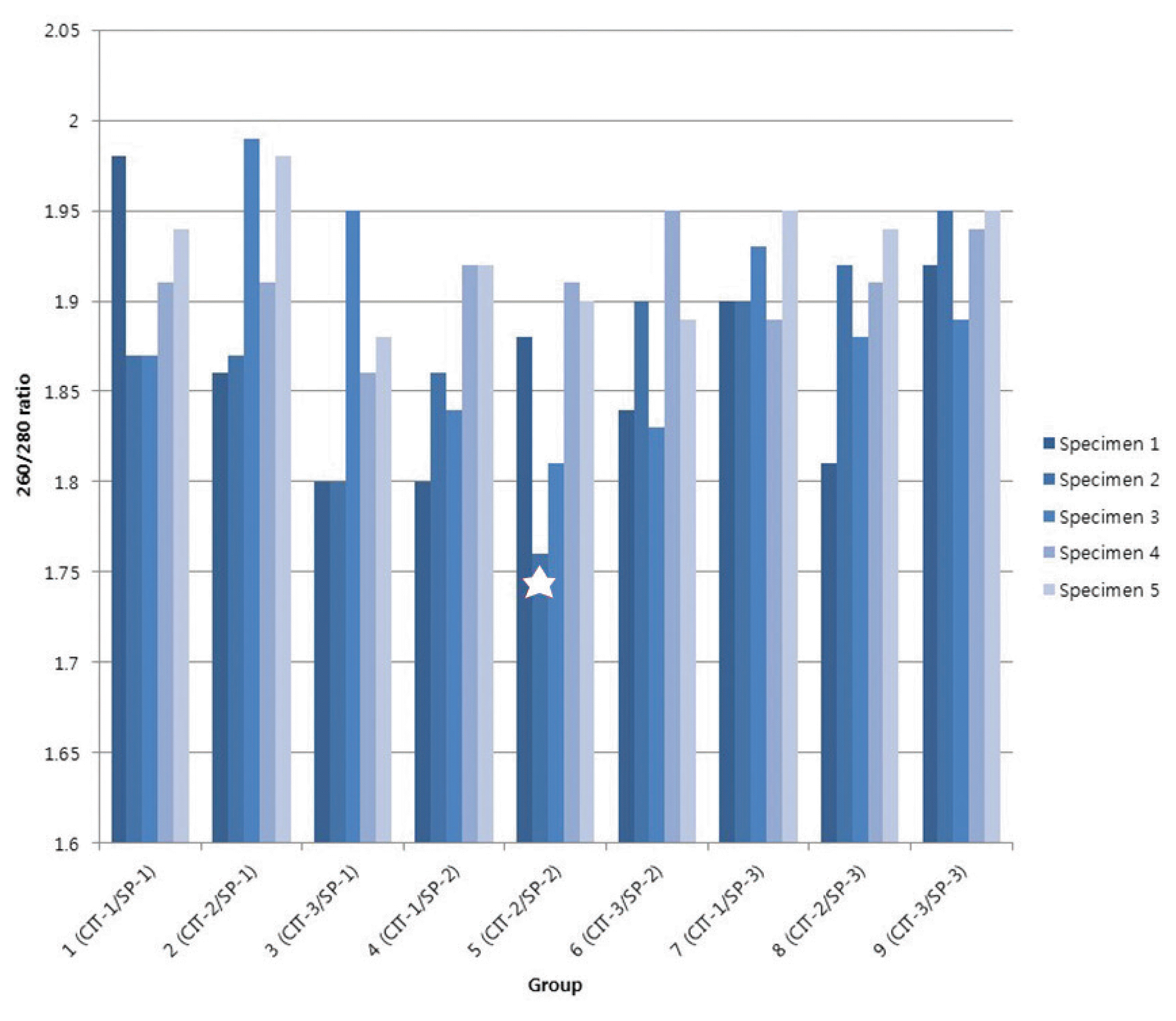 | Fig. 1DNA purity (260/280 ratio) in 45 biobanked fresh frozen colorectal cancer tissues. A single specimen showed out of DNA purity range with 1.76 of 260/280 ratio (★).
CIT = cold ischemia time; SP = storage period; CIT-1 = less than 30 minutes; CIT-2 = 31–45 minutes; CIT-3 = 46–60 minutes; SP-1 = less than 1 year; SP-2 = 2–3 years; SP-3 = 4–5 years.
|
DNA integrity of different CIT and SP
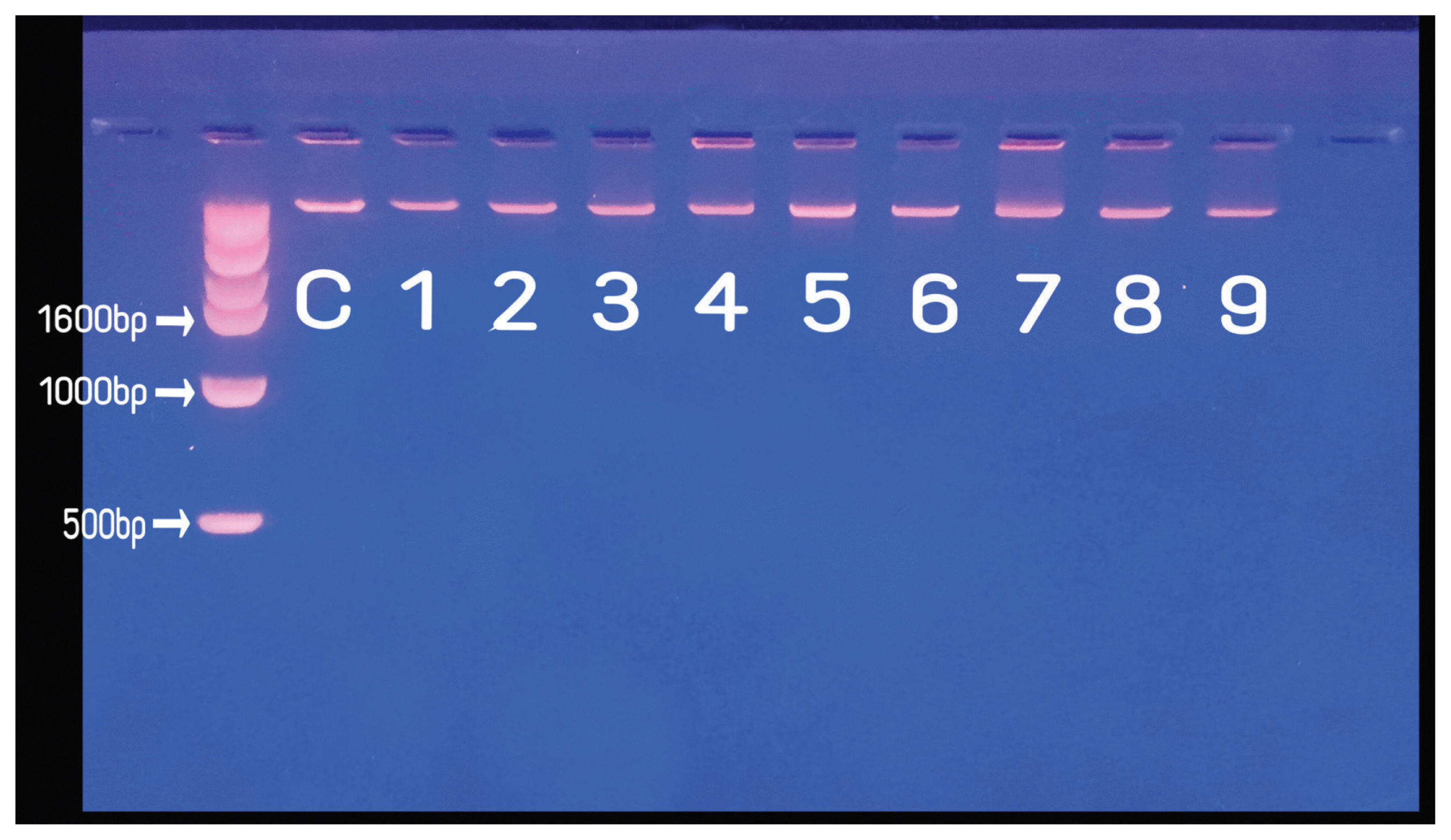
All DNA specimens appeared as a compact, high-molecular-weight band with no or scanty low-molecular-weight smears in the gel electrophoresis. According to our results, 100% of extracted DNA specimens were of good quality and no significant difference was found in DNA quality of different CIT. DNA band from agarose gel results for group 1, 2, and 3 representing less than 1 year of SP appeared to be clearer than groups 4–9 representing more than 1 year of SP, but which showed no significant difference (Fig. 2).
KARS mutation status
The DNA quality was also assessed by PCR amplification of KRAS gene to evaluate practical usability of DNA from a biobanked tissue for molecular study. KRAS mutation status of all 45 biobanked frozen tissues that was performed at the time of current study was compared with the results of corresponding FFPE tissues that was initially performed at the time of pathologic diagnosis. The concordance of KRAS mutation status between all 45 paired biobanked frozen tissues and FFPE tissues was 100%, with 14 paired specimens showing the same KRAS mutations (Fig. 3). KRAS mutation was found in 31.1% (14/45) of CRC. The most common mutation locations were in codon 12 (64.3%, 9/14) and codon 13 (28.6%, 4/14). One specimen (7.1%, 1/14) had 2 synchronous mutations in codon 12 and codon 13. According to the PCR results, all the Ct values were stable at different CIT or SP.
Go to : 
DISCUSSION
Biobanking involves the collection, processing, transport, storage, and retrieval of biospecimens for future researches.2 Biobanked specimens are considered as invaluable future resources to carry out molecular biology, cell biology, genetics, transcriptomics, genomics and proteomics research in order to explore new standards of tumor classification, diagnosis, treatment, and prognosis.8
However, the quality of DNA or RNA expression in biobanked specimens is dependent on multiple preanalytical variables such as tissue type, intrinsic patient factors, warm IT (i.e., extraction of the specimen after ligation of the large vessels), CIT, fixation method, subsequent transport, storage condition, and SP of specimens.14 While tissue type and intrinsic patient factors cannot be modified, other factors such as CIT and SP can be controlled. Although several studies reported the impact of CIT and SP on DNA integrity of variable biobanked tissue, it is not well documented. 5–10,15 For CRC tissue, one study reported that a critical time point for tissue handling appeared to be 1 hour at room temperature,15 whereas other studies have shown that RNA remained stable when kept at room temperature and on ice for up to 4 hours.16 A recent review specifically addressing the effect of CIT on RNA stability concluded that in most of the studies, only minimal changes (≤ 10%) in the RNA integrity number in CRC tissues were observed during a CIT of 1 – 6 hours.9 Guerrera et al.17 reported that patient-derived xenografts engraftment rate was adversely affected by prolonged CITs (> 10 hours). Regarding SP, extended cryogenic storage beyond 2–11 years remained a viable option for maintaining the high quality of various tissue specimens in biobanks.10 Long-term storage of biobanked CRC specimens did not negatively influence DNA and RNA qualities, for 2 years14 and up to 40 months.16 In the present study, we found no significant difference in DNA purity and integrity of different CITs (up to 1 hour) and SPs (at least 5 years) similar to other studies. Apparently, it is evident that storage conditions of our biobank are suitable for long-term specimen preservation with high DNA quality.
To determine the suitability of biobanked frozen tissue for biomarker research according to various CITs or SPs, the mutation status of KRAS was investigated in all paired FFPE and biobanked frozen tissues. KRAS, a well-known signaling molecule in the EGFR pathway, has been recognized as one of the most frequently mutated oncogenes in CRC.11 Mutations in codons 12/13 of KRAS exon 2 are associated with reduced benefit from anti-EGFR antibody treatment for metastatic CRC.18 Our study showed 100% concordance of KRAS mutation in all 45 paired FFPE and biobanked frozen tissues with variable CITs and SPs, thereby suggesting that biobanked tissues with 1-hour CIT and 5 years SP are suitable for biomarker research. In the near future, significant advancements in tissue-sequencing platforms will be made to understand molecular characterization and discover novel biomarkers for predicting treatment response for CRC patients. Consequently, there is a need to reconsider the utility of specimens stored beyond a certain timeframe. In this context, high-quality biobanked frozen tissue would hold practical significance with utilization as alternative biospecimens to investigate future relevant molecular signatures, when archival FFPE block is not available.
Despite the advantages of FFPE tissues including abundance and availability, connection to rich clinical data, and association with patient outcomes, 19 another issue involves C > T transition artifacts associated with FFPE-derived DNA as a result of the addition of adenine instead of guanosine due to deamination.20 Previous studies demonstrated that pairwise analysis of DNA exome-sequencing data showed concordance for 70 – 80% of variants in biobanked frozen tissue and FFPE specimens stored for fewer than 3 years21 and next-generation sequencing results from biobanked frozen specimen were more consistent than FFPE specimens.22 Although RNA-sequencing data showed a high correlation between expression profiles in biobanked frozen tissue/FFPE irrespective of storage time (up to 244 months) and tissue type, there exist discordances between RNA-sequencing results from FFPE and biobanked frozen tissue.21 In addition, high failure rate in FFPE specimens to achieve sufficient quality RNA for RNA-sequencing has been demonstrated.19 FFPE tissue processing and specimen storage are known to result in highly degraded and chemically modified RNA, which limits gene detection and introduces sequencing artifacts.19,23 Unlike FFPE tissue, the DNA and RNA from frozen biospecimens are generally of high molecular weight and without cross-linking, which are ideal for a wide variety of purposes such as whole-genome amplification, whole genome sequencing, and cDNA microarray analyses, when the tissue biobanking is precisely controlled under the SOP.
In this context, all tissue specimens for biobanking must be precisely handled under the SOP. SOP addresses multiple factors including collection, preparation, transport, storage, management, instrumentation, facilities, quality control, and utilization of fresh tissue for standardized specimen consistency, accuracy, and quality.24 The SOP is also needed to avoid high costs and the disposal of valuable biospecimens. Furthermore, researchers carry out large-scale, multicenter studies and SOP is required to standardize different collections and for quality management of high-quality specimens within different institutions to assure good scientific practice.22 Since the Korea Biobank Project started in 2008,25 many medical college- or hospital-based biobanks have been in operation in Korea. As the SOP for the biobank of Korea was developed quite recently, 25,26 there is lot of unexploited potential for scientific questions. We hypothesize that this study would partly contribute towards standardization of collection, storage, and quality control of fresh frozen tissues in Korea biobank.
There are a few limitations to our study. Firstly, this study did not represent the full diversity of the preanalytical tissue handling process in tissue biobanking and could not perfectly exclude other factors having a major impact on DNA quality. However, we were able to analyze the most important modifiable factors such as CIT and SP and show actual specimen handling data as per institutional review board-approved SOP. Another limitation is that the present study only provided the influence of CIT and SP on DNA quality. As ischemia has a more significant influence on RNA integrity, results should thus be confirmed based on analysis of RNA according to preanalytical variables for performing emerging systematic techniques such as next-generation RNA sequencing. The third limitation is that the present study could not provide data on cold ischemia tolerance according to specific tissue types, because the subject studied was only CRC tissue. Further studies involving various tissues are necessary to elucidate whether other tumors demonstrate similar quality results and feasibility of collection protocols for specific tissue types. Finally, our biobank has been established only for a short period (presently up to six years), further studies with increased CIT and SP points are necessitated to validate this observation and find optimal CIT and SP.
In conclusion, we found no significant difference in DNA purity and integrity of different CITs (up to 1 hour) and SPs (at least 5 years) in biobanked CRC specimens. Moreover, the storage conditions of our biobank are considered suitable for biomarker research involving high DNA quality. The findings of this study have practical implications that could affect biospecimen collection and banking guidelines.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download