Abstract
Pancreatic ductal adenocarcinoma is a lethal cancer type that is associated with multiple gene mutations in somatic cells. Genetically engineered mouse is hardly applicable for developing a pancreatic cancer model, and the xenograft model poses a limitation in the reflection of early stage pancreatic cancer. Thus, in vivo somatic cell gene engineering with clustered regularly interspaced short palindromic repeats is drawing increasing attention for generating an animal model of pancreatic cancer. In this study, we selected Kras, Trp53, Ink4a, Smad4, and Brca2 as target genes, and applied Campylobacter jejuni Cas9 (CjCas9) and Streptococcus pyogens Cas9 (SpCas9) for developing pancreatic cancer using adeno associated virus (AAV) transduction. After confirming multifocal and diffuse transduction of AAV2, we generated SpCas9 overexpression mice, which exhibited high double-strand DNA breakage (DSB) in target genes and pancreatic intraepithelial neoplasia (PanIN) lesions with two AAV transductions; however, wild-type (WT) mice with three AAV transductions did not develop PanIN. Furthermore, small-sized Cjcas9 was applied to WT mice with two AAV system, which, in addition, developed high extensive DSB and PanIN lesions. Histological changes and expression of cancer markers such as Ki67, cytokeratin, Mucin5a, alpha smooth muscle actin in duct and islet cells were observed. In addition, the study revealed several findings such as 1) multiple DSB potential of AAV-CjCas9, 2) periductal lymphocyte infiltration, 3) multi-focal cancer marker expression, and 4) requirement of > 12 months for initiation of PanIN in AAV mediated targeting. In this study, we present a useful tool for in vivo cancer modeling that would be applicable for other disease models as well.
Go to : 
References
3. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA. Australian Pancreatic Cancer Genome InitiativeGill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015; 518:495–501.

4. Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Klöppel G, Longnecker DS, Lüttges J, Offerhaus GJ. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001; 25:579–586.
5. Behrens D, Walther W, Fichtner I. Pancreatic cancer models for translational research. Pharmacol Ther. 2017; 173:146–158.

6. Herreros-Villanueva M, Hijona E, Cosme A, Bujanda L. Mouse models of pancreatic cancer. World J Gastroenterol. 2012; 18:1286–1294.

7. Gopinathan A, Morton JP, Jodrell DI, Sansom OJ. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech. 2015; 8:1185–1200.

8. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821.

9. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823.

10. Feng W, Jasin M. Homologous recombination and replication fork protection: BRCA2 and more! Cold Spring Harb Symp Quant Biol. 2017; 82:329–338.
11. Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, Jacks T. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014; 516:428–431.

12. Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014; 159:440–455.

13. Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014; 514:380–384.

14. Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015; 6:7391.

15. Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, Brady JJ, Yang D, Grüner BM, Chuang CH, Caswell DR, Zeng H, Chu P, Kim GE, Carpizo DR, Kim SK, Winslow MM. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015; 29:1576–1585.

16. Maresch R, Mueller S, Veltkamp C, Öllinger R, Friedrich M, Heid I, Steiger K, Weber J, Engleitner T, Barenboim M, Klein S, Louzada S, Banerjee R, Strong A, Stauber T, Gross N, Geumann U, Lange S, Ringelhan M, Varela I, Unger K, Yang F, Schmid RM, Vassiliou GS, Braren R, Schneider G, Heikenwalder M, Bradley A, Saur D, Rad R. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. 2016; 7:10770.

17. Ideno N, Yamaguchi H, Okumura T, Huang J, Brun MJ, Ho ML, Suh J, Gupta S, Maitra A, Ghosh B. A pipeline for rapidly generating genetically engineered mouse models of pancreatic cancer using in vivo CRISPR-Cas9-mediated somatic recombination. Lab Invest. 2019; 99:1233–1244.

18. Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel). 2017; 9:E42.

19. Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017; 8:14500.
20. Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D, Meng S, Zhang X, Hu S, Tian Y. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep. 2014; 4:6420.

21. Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, Stoner R. Inference of CRISPR edits from sanger trace data. bioRxivorg. 2019. Epub ahead of print.DOI: doi: 10.1101/251082.

22. Wang D, Tai PW, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019; 18:358–378.

23. Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016; 351:400–403.

24. Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015; 33:102–106.

25. Brembeck FH, Schreiber FS, Deramaudt TB, Craig L, Rhoades B, Swain G, Grippo P, Stoffers DA, Silberg DG, Rustgi AK. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 2003; 63:2005–2009.
26. Fujita H, Ohuchida K, Mizumoto K, Nakata K, Yu J, Kayashima T, Cui L, Manabe T, Ohtsuka T, Tanaka M. Alpha-smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas. 2010; 39:1254–1262.

27. Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013; 10:607–620.

28. Panzuto F, Cicchese N, Partelli S, Rinzivillo M, Capurso G, Merola E, Manzoni M, Pucci E, Iannicelli E, Pilozzi E, Rossi M, Doglioni C, Falconi M, Delle Fave G. Impact of Ki67 re-assessment at time of disease progression in patients with pancreatic neuroendocrine neoplasms. PLoS One. 2017; 12:e0179445.

29. Doiron B, DeFronzo RA. A novel experimental model for human mixed acinar-ductal pancreatic cancer. Carcinogenesis. 2018; 39:180–190.

30. Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014; 15:445–451.

31. Quirin KA, Kwon JJ, Alioufi A, Factora T, Temm CJ, Jacobsen M, Sandusky GE, Shontz K, Chicoine LG, Clark KR, Mendell JT, Korc M, Kota J. Safety and efficacy of AAV retrograde pancreatic ductal gene delivery in normal and pancreatic cancer mice. Mol Ther Methods Clin Dev. 2017; 8:8–20.

32. Ferreira RM, Sancho R, Messal HA, Nye E, Spencer-Dene B, Stone RK, Stamp G, Rosewell I, Quaglia A, Behrens A. Duct- and acinar-derived pancreatic ductal adenocarcinomas show distinct tumor progression and marker expression. Cell Reports. 2017; 21:966–978.

34. Schachtschneider KM, Schwind RM, Newson J, Kinachtchouk N, Rizko M, Mendoza-Elias N, Grippo P, Principe DR, Park A, Overgaard NH, Jungersen G, Garcia KD, Maker AV, Rund LA, Ozer H, Gaba RC, Schook LB. The oncopig cancer model: an innovative large animal translational oncology platform. Front Oncol. 2017; 7:190.

35. Cicenas J, Tamosaitis L, Kvederaviciute K, Tarvydas R, Staniute G, Kalyan K, Meskinyte-Kausiliene E, Stankevicius V, Valius M. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol. 2017; 34:26.

36. Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Shah RR, Shah A, Ling D, Growe J, Pink M, Rohde E, Wood KM, Salomon WE, Harrington WF, Dombrowski C, Strapps WR, Chang Y, Morrissey DV. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Reports. 2018; 22:2227–2235.

Go to : 
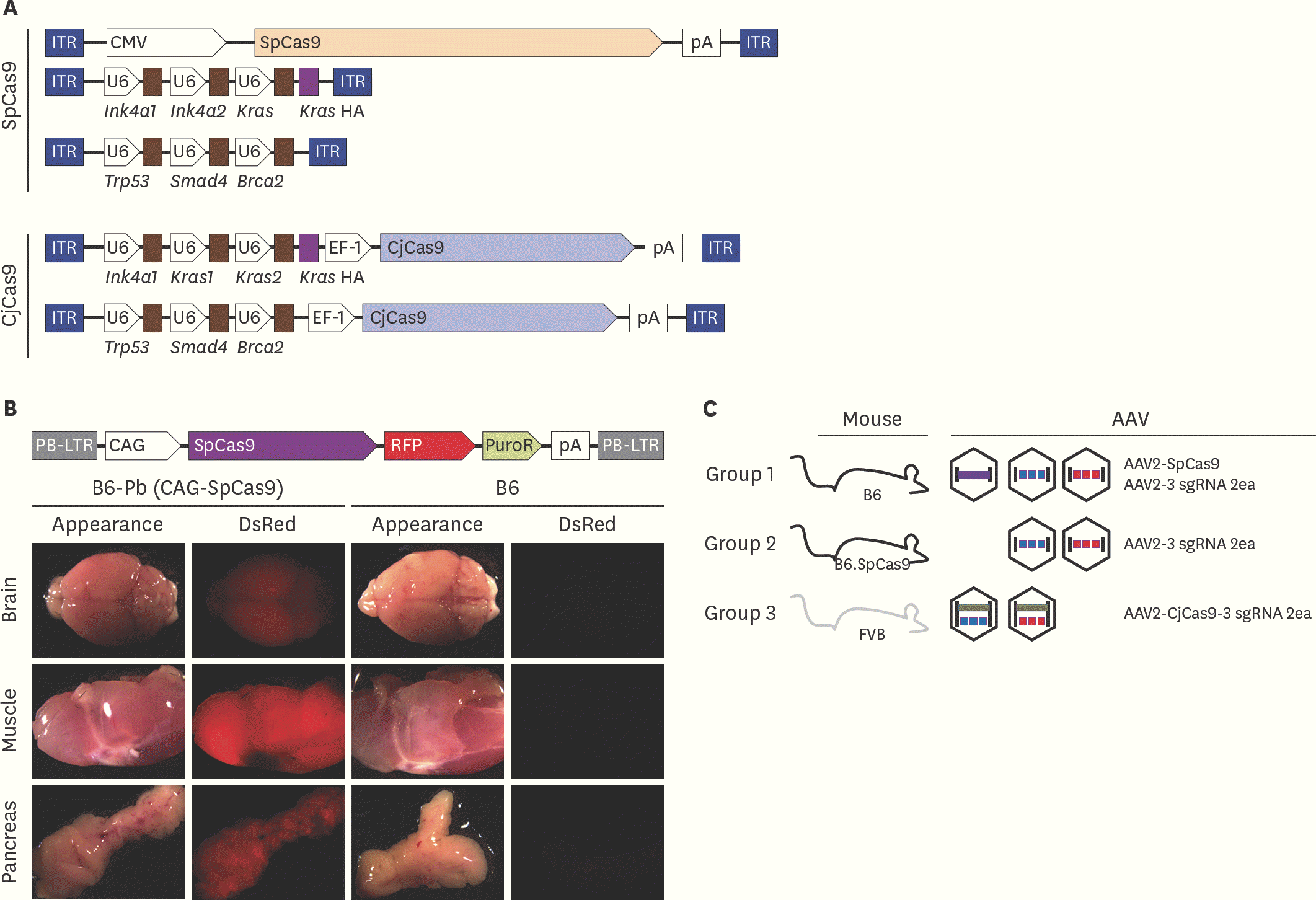 | Fig. 1.Brief schematic of in vivo gene targeting with CRISPR/Cas9 (A) Vector maps of AAV-SpCas9, AAV-3 sgRNA, and AAV- CjCas9-all-in-one used in this study. Five different rAAV vectors were prepared by synthesis and cloning. Blue box: ITR sequence from AAV2, arrow box: promoter, brown box: sgRNA sequence. (B) SpCas9 overexpression mouse generation with PiggyBac transposon system. pCAG-SpCas9-RFP-pA insert was incorporated into host genome by transposase. Gene integration confirmed by RFP expression under stereoscope microscope. (C) Experimental group of in vivo gene targeting for pancreatic cancer development. WT C57BL/6 mice were injected with three AAVs; AAV2-SpCas9, AAV2-3 sgRNA (Kras-Ink4a1-In4a2-HA) and AAV2-3 sgRNA (Trp53-Smad4-Brca2) into common bile duct in group 1. Instead of AAV2-SpCas9 injection, SpCas9 overexpression mice were applied in group 2. WT FVB mice and all-in-one type two AAVs (AAV2-CjCas9-sgRNA for Kras, Ink4a-HA and AAV2-CjCas9-sgRNA for Trp53, Smad4, Brca2) were injected in group 3. Detailed information of each target is provided in Supplementary Fig. 1 and Supplementary Table 1. AAV, adeno associated virus; SpCas9, Streptococcus pyogens Cas9; CjCas9, Campylobacter jejuni Cas9; sgRNA, single guide RNA; CRISPR, clustered regularly interspaced short palindromic repeats; rAAV, recombinant adeno associated virus; WT, wild-type. |
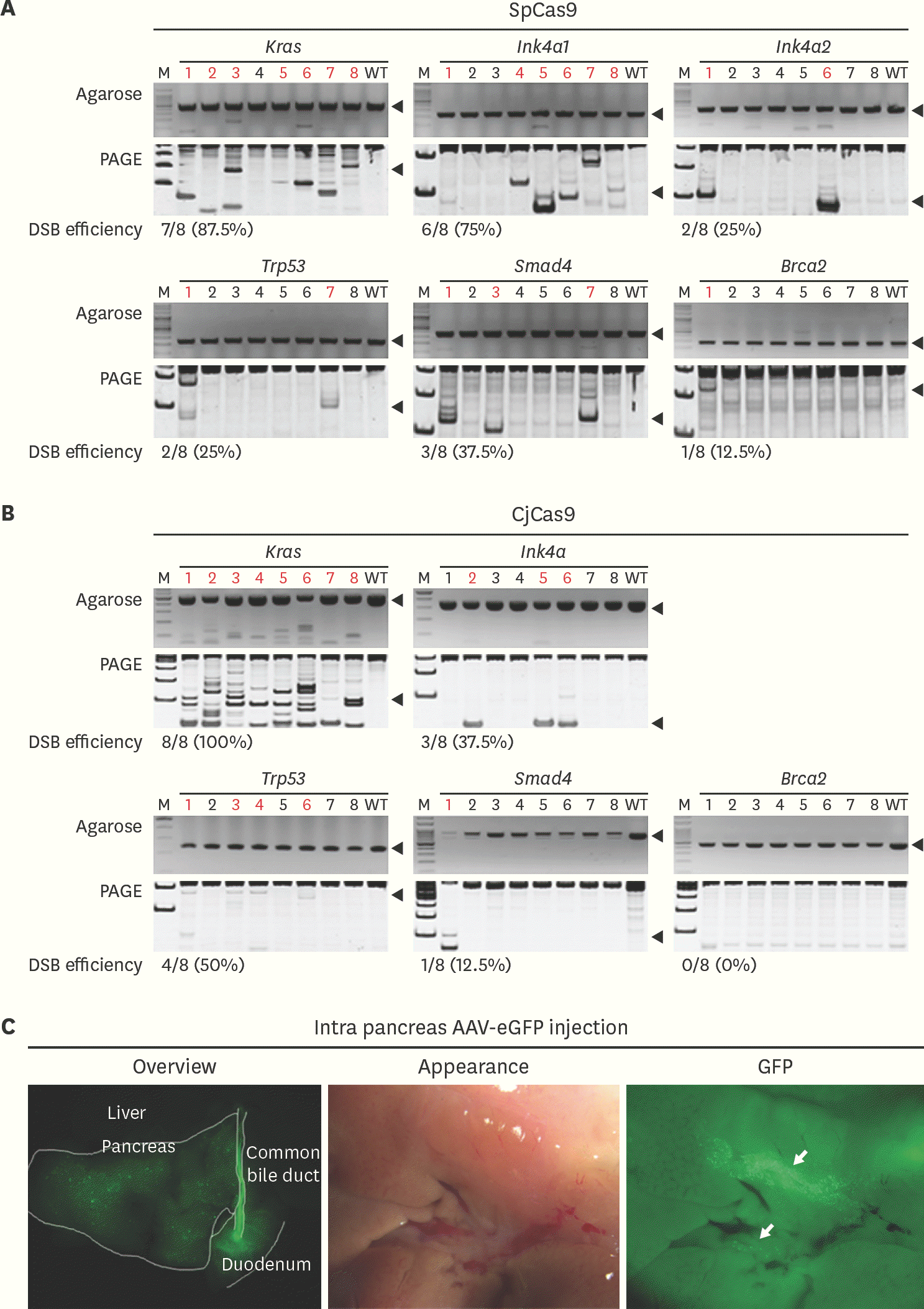 | Fig. 2.Evaluation of DSB efficiency and AAV2 tropism in pancreas (A) and (B). Cas9 messenger RNA and 3 single guide RNAs plasmid were co-injected into 1 cell stage embryos and DSB was evaluated by hetero-duplex formation and SDS-PAGE gel. Red letters indicate DSB and black arrow indicates the target size of each polymerase chain reaction reaction. DSB efficiency presented as number of embryos with DSB/number of total embryos used (%). (C) After 3 weeks of AAV2-eGFP injection into pancreas, GFP expression analyzed with stereoscopic microscope. White guideline distinguishes each of the organs and white arrows indicates spots for GFP expression. Statistical analysis was performed using Student's t-test. SpCas9, Streptococcus pyogens Cas9; CjCas9, Campylobacter jejuni Cas9; DSB, double-strand DNA breakage; SDS-PAGE, sodium dodecyl sulfate– polyacrylamide gel electrophoresis; AAV, adeno associated virus; eGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; WT, wild-type. |
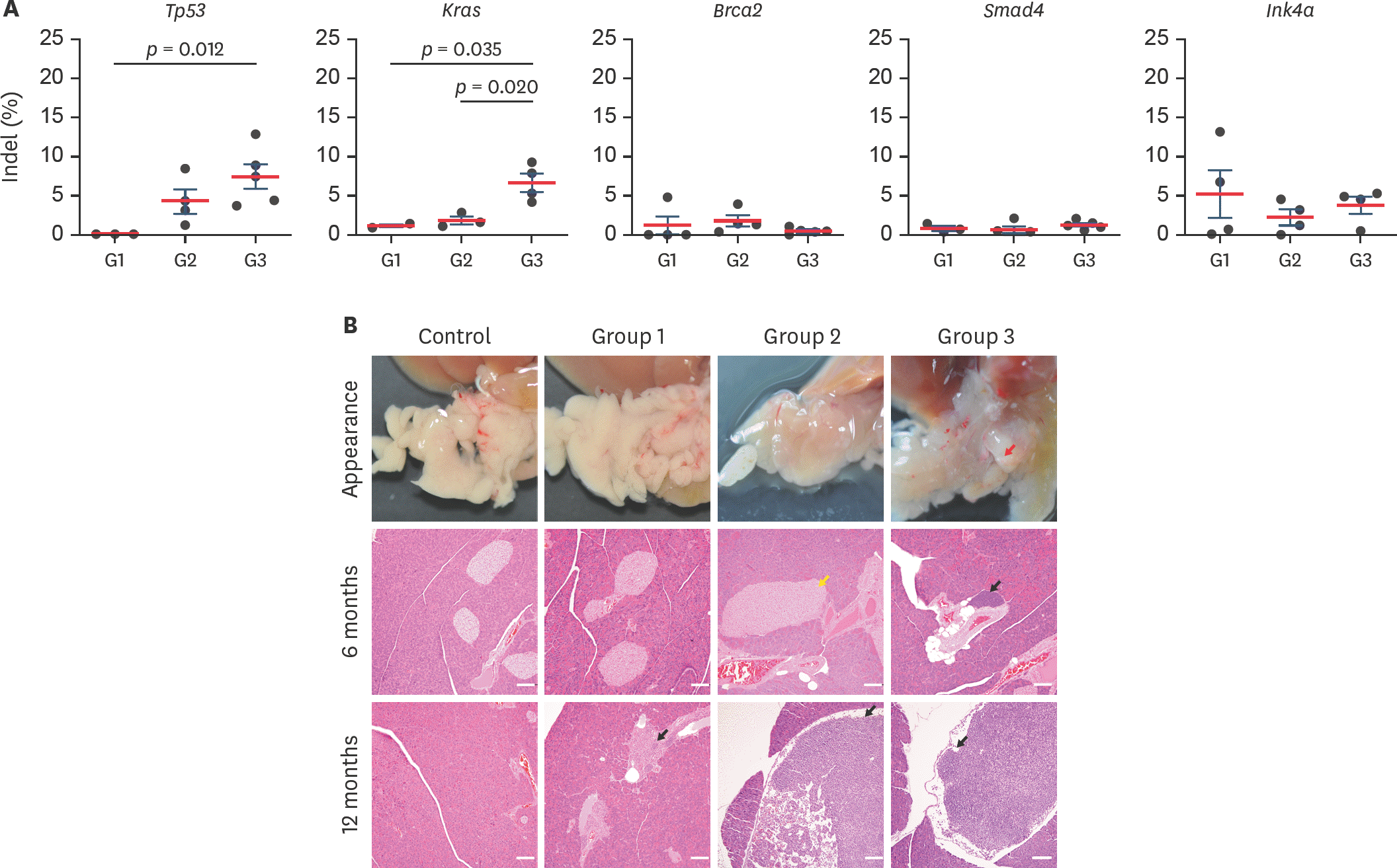 | Fig. 3.DSB frequency analysis and macroscopic examination (A) Indel frequency of target genes was analyzed with Sanger sequencing and algorithm of Synthego ICE tool. Each dot indicates DSB rate from each mice. Statistical analysis was performed with Student's t-test, and p < 0.05 indicated significantly different. (B) Appearance and microscopic analysis on pancreas. Red arrow: abnormal lesion, yellow arrow: islet hyperplasia, black arrow: periductal lymphocyte infiltration and mesentery lymph node, white bar, 200 µm. DSB, double-strand DNA breakage. |
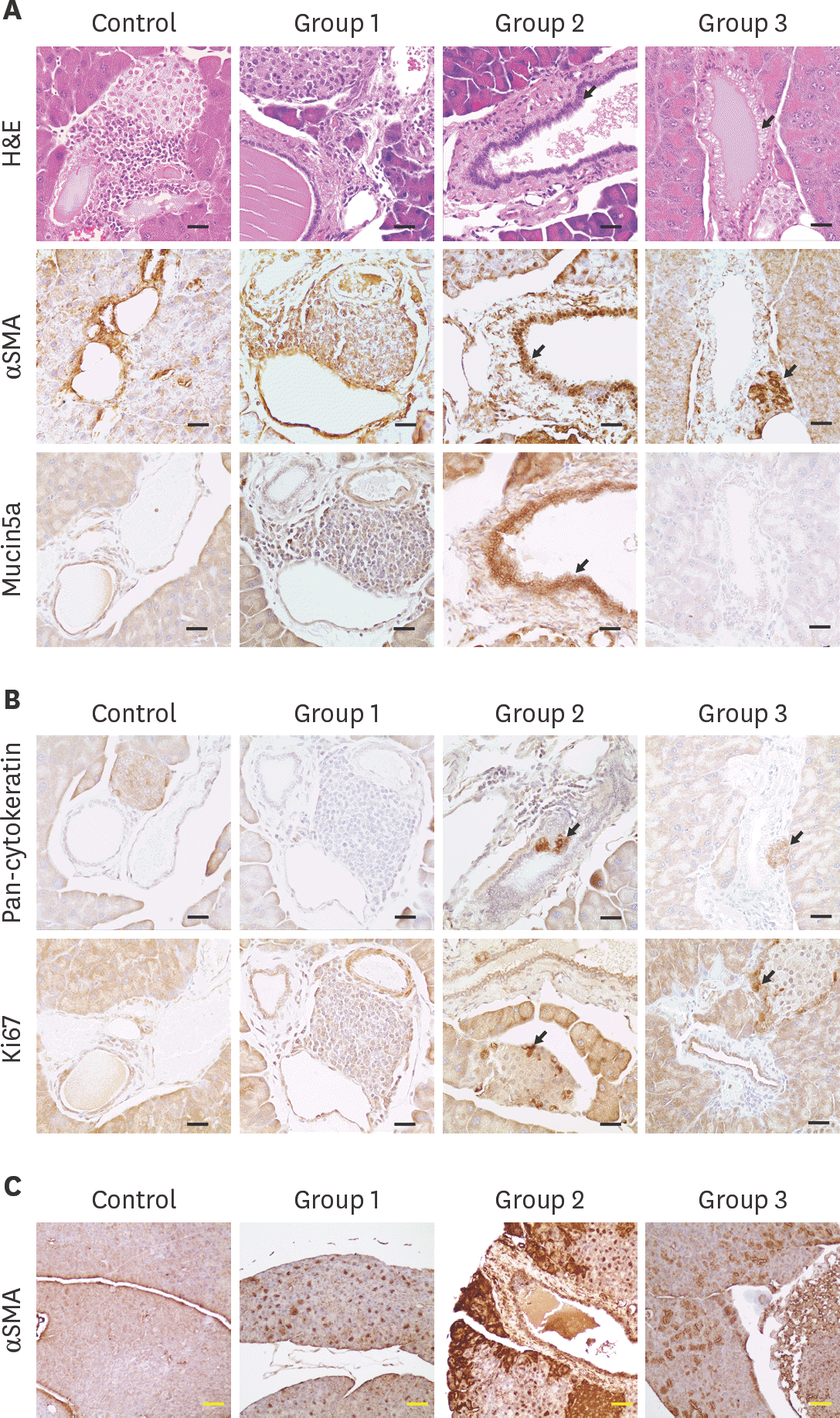 | Fig. 4.Histological and IHC analysis for evaluation of pancreatic cancer development (A) and (B) H&E stain and IHC for pan-cytokeratin, α SMA, Mucin5a, and Ki67 was conducted with formalin fixed pancreatic tissues. Black arrow: PanIN lesion or cancer marker expression. Black scale bar: 50 µm (C) IHC for α SMA marker. Yellow scale bar, 100 µm. IHC, immunohistochemistry; α SMA, alpha smooth muscle actin; H&E, hematoxylin and eosin; PanIN, pancreatic intraepithelial neoplasia. |
Table 1.
Summary of in vivo gene targeting for developing pancreatic cancer




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download