Abstract
Rabid raccoon dogs (Nyctereutes procyonoides koreensis) have been responsible for animal rabies in South Korea since the 1990s. A recombinant rabies vaccine strain, designated as ERAGS, was constructed for use as a bait vaccine. Therefore, new means of differentiating ERAGS from other rabies virus (RABV) strains will be required in biological manufacturing and diagnostic service centers. In this study, we designed two specific primer sets for differentiation between ERAGS and other RABVs based on mutation in the RABV glycoprotein gene. Polymerase chain reaction analysis of the glycoprotein gene revealed two DNA bands of 383 bp and 583 bp in the ERAGS strain but a single DNA band of 383 bp in the field strains. The detection limits of multiplex reverse transcription polymerase chain reaction (RT-PCR) were 80 and 8 FAID50/reaction for the ERAGS and Evelyn-Rokitnicki-Abelseth strains, respectively. No cross-reactions were detected in the non-RABV reference viruses, including canine distemper virus, parvovirus, canine adenovirus type 1 and 2, and parainfluenza virus. The results of multiplex RT-PCR were 100% consistent with those of the fluorescent antibody test. Therefore, one-step multiplex RT-PCR is likely useful for differentiation between RABVs with and those without mutation at position 333 of the RABV glycoprotein gene.
Go to : 
References
1. Walker PJ, Blasdell KR, Calisher CH, Dietzgen RG, Kondo H, Kurath G, Longdon B, Stone DM, Tesh RB, Tordo N, Vasilakis N, Whitfield AE. Ictv Report Consortium. ICTV virus taxonomy profile: rhabdoviridade. J Gen Virol. 2018; 99:447–448.
2. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. Field Virology. Lippincott Williams & Wilkins;Philadelphia: 2001.
3. Morimoto K, Hooper DC, Spitsin S, Koprowski H, Dietzschold B. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J Virol. 1999; 73:510–518.

4. Seif I, Coulon P, Rollin PE, Flamand A. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J Virol. 1985; 53:926–934.

5. Faber M, Faber ML, Papaneri A, Bette M, Weihe E, Dietzschold B, Schnell MJ. A single amino acid change in rabies virus glycoprotein increases virus spread and enhances virus pathogenicity. J Virol. 2005; 79:14141–14148.

6. Huang Y, Tang Q, Rayner S, Gong K, Song B, Liang GD. Pathogenicity of rabies viruses isolated in China: two fixed strains and a street strain. Biomed Environ Sci. 2013; 26:552–561.
7. Lawson KF, Crawley JF. The ERA strain of rabies vaccine. Can J Comp Med. 1972; 36:339–344.
8. Tao L, Ge J, Wang X, Zhai H, Hua T, Zhao B, Kong D, Yang C, Chen H, Bu Z. Molecular basis of neurovirulence of flury rabies virus vaccine strains: importance of the polymerase and the glycoprotein R333Q mutation. J Virol. 2010; 84:8926–8936.

9. Le Blois H, Tuffereau C, Blancou J, Artois M, Aubert A, Flamand A. Oral immunization of foxes with avirulent rabies virus mutants. Vet Microbiol. 1990; 23:259–266.

10. Sato G, Kobayashi Y, Motizuki N, Hirano S, Itou T, Cunha EM, Ito FH, Sakai T. A unique substitution at position 333 on the glycoprotein of rabies virus street strains isolated from non-hematophagous bats in Brazil. Virus Genes. 2009; 38:74–79.

11. Madhusudana SN, Tripathi KK. Oral infectivity of street and fixed rabies virus strains in laboratory animals. Indian J Exp Biol. 1990; 28:497–499.
12. Yang DK, Kim HH, Lee KW, Song JY. The present and future of rabies vaccine in animals. Clin Exp Vaccine Res. 2013; 2:19–25.

13. World Organization for Animal Health (OIE). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 7th ed.OIE;Paris: 2012.
14. Yang DK, Kim HH, Choi SS, Kim JT, Lee KB, Lee SH, Cho IS. Safety and immunogenicity of recombinant rabies virus (ERAGS) in mice and raccoon dogs. Clin Exp Vaccine Res. 2016; 5:159–168.

15. Franka R, Wallace R. Rabies diagnosis and surveillance in animals in the era of rabies elimination. Rev Sci Tech. 2018; 37:359–370.

16. Nakagawa K, Nakagawa K, Omatsu T, Katayama Y, Oba M, Mitake H, Okada K, Yamaoka S, Takashima Y, Masatani T, Okadera K, Ito N, Mizutani T, Sugiyama M. Generation of a novel live rabies vaccine strain with a high level of safety by introducing attenuating mutations in the nucleoprotein and glycoprotein. Vaccine. 2017; 35:5622–5628.

17. Tian Q, Wang Y, Zhang Q, Luo J, Mei M, Luo Y, Guo X. Rescue of a wild-type rabies virus from cloned cDNA and assessment of the proliferative capacity of recombinant viruses. Virus Genes. 2017; 53:573–583.

18. Deubellbeiss A, Zahno ML, Zanoni M, Bruegger D, Zanoni R. Real-time RT-PCR for the detection of Lyssavirus species. J Vet Med. 2014; 2014:476091.
Go to : 
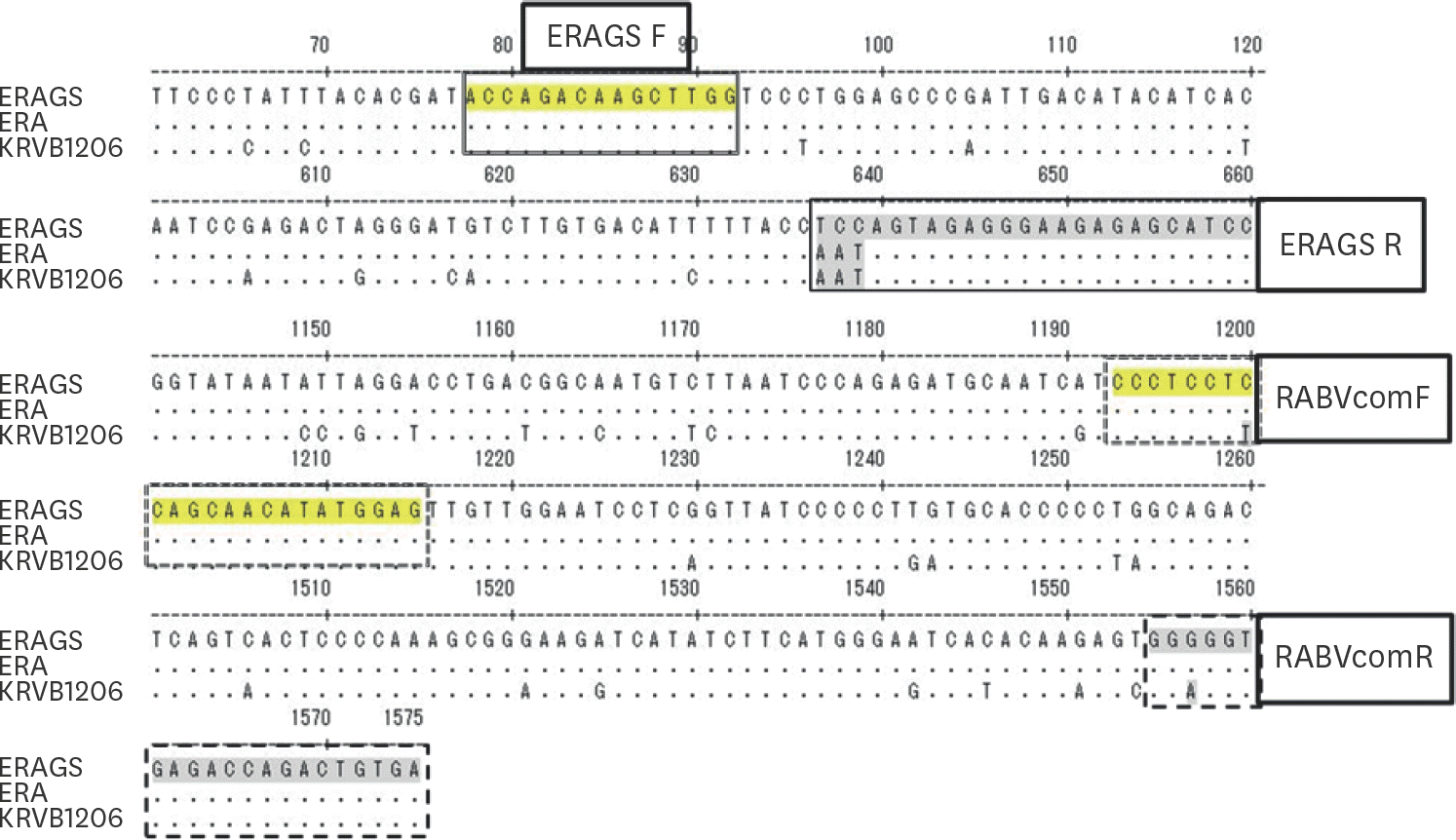 | Fig. 1.RABV G gene sites targeted by the primers used for differential RT-PCR. The first three nucleotides (TCC) of the ERAGS reverse primer, corresponding to codon 333 of the G gene, were not identical to those (AAT) of the Evelyn-Rokitnicki-Abelseth strain or KRVB1206 isolate. |
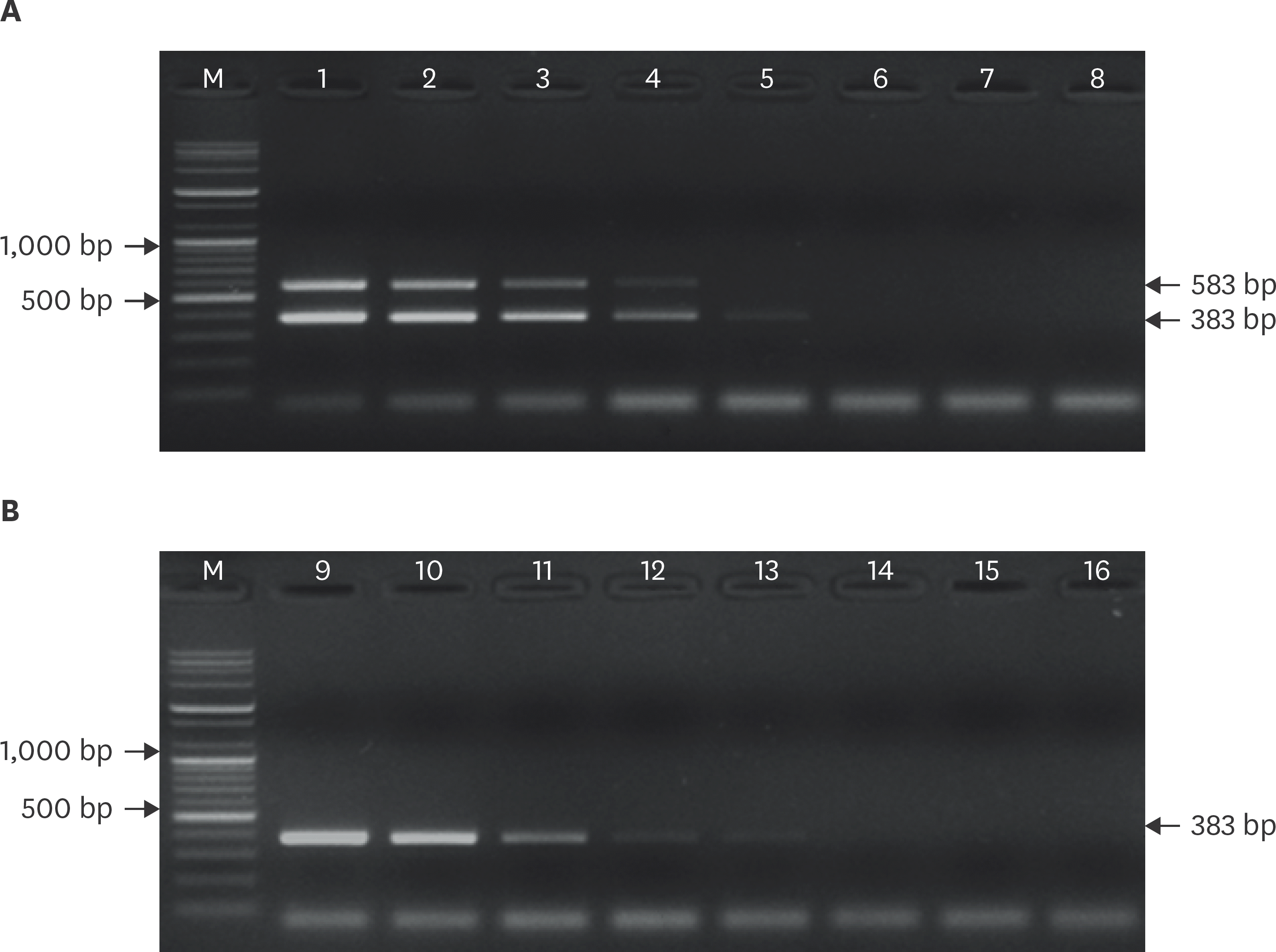 | Fig. 2.Sensitivity of the specific primer sets used for differential detection of ERAGS and ERA strains. M, 100 bp DNA ladder; lanes 1–8, ERAGS strain 100–10-7 (A). M, 100 bp DNA ladder; lanes 9–16, ERA strain 100–10-7 (B). ERA, Evelyn-Rokitnicki-Abelseth. |
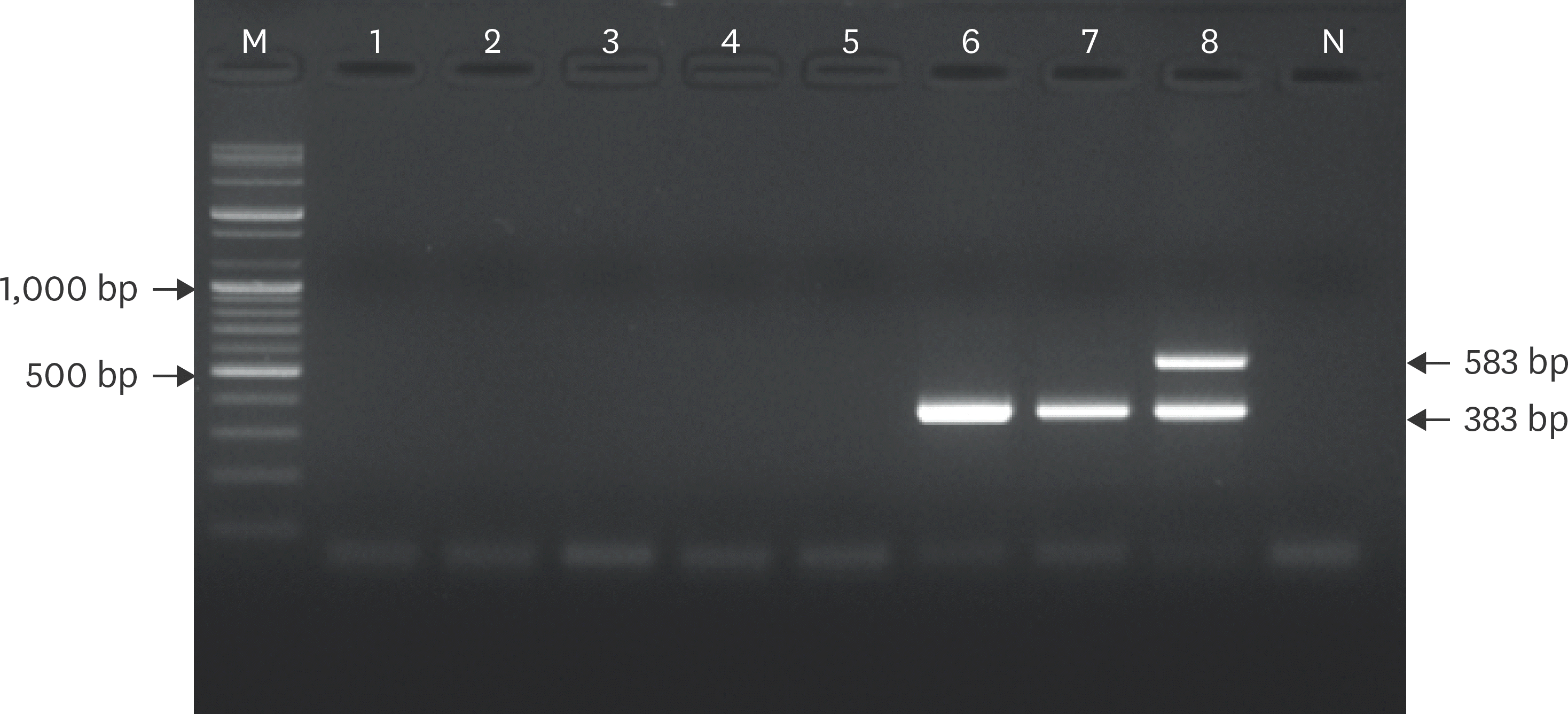 | Fig. 3.Specificity of differential RABV RT-PCR using two primer sets. RT-PCR detected only RABVs, and no positive signals were obtained for the five canine viral pathogens. M, 100 bp DNA ladder; lane 1, canine distemper virus; lane 2, canine parainfluenza virus; lane 3, canine adenovirus type 1; lane 4, canine adenovirus type 2; lane 5, canine parvovirus; lane 6, CVS11; lane 7, Evelyn-Rokitnicki-Abelseth strain; lane 8, ERAGS strain; lane N, negative control (water only). RT-PCR, reverse transcription polymerase chain reaction. |
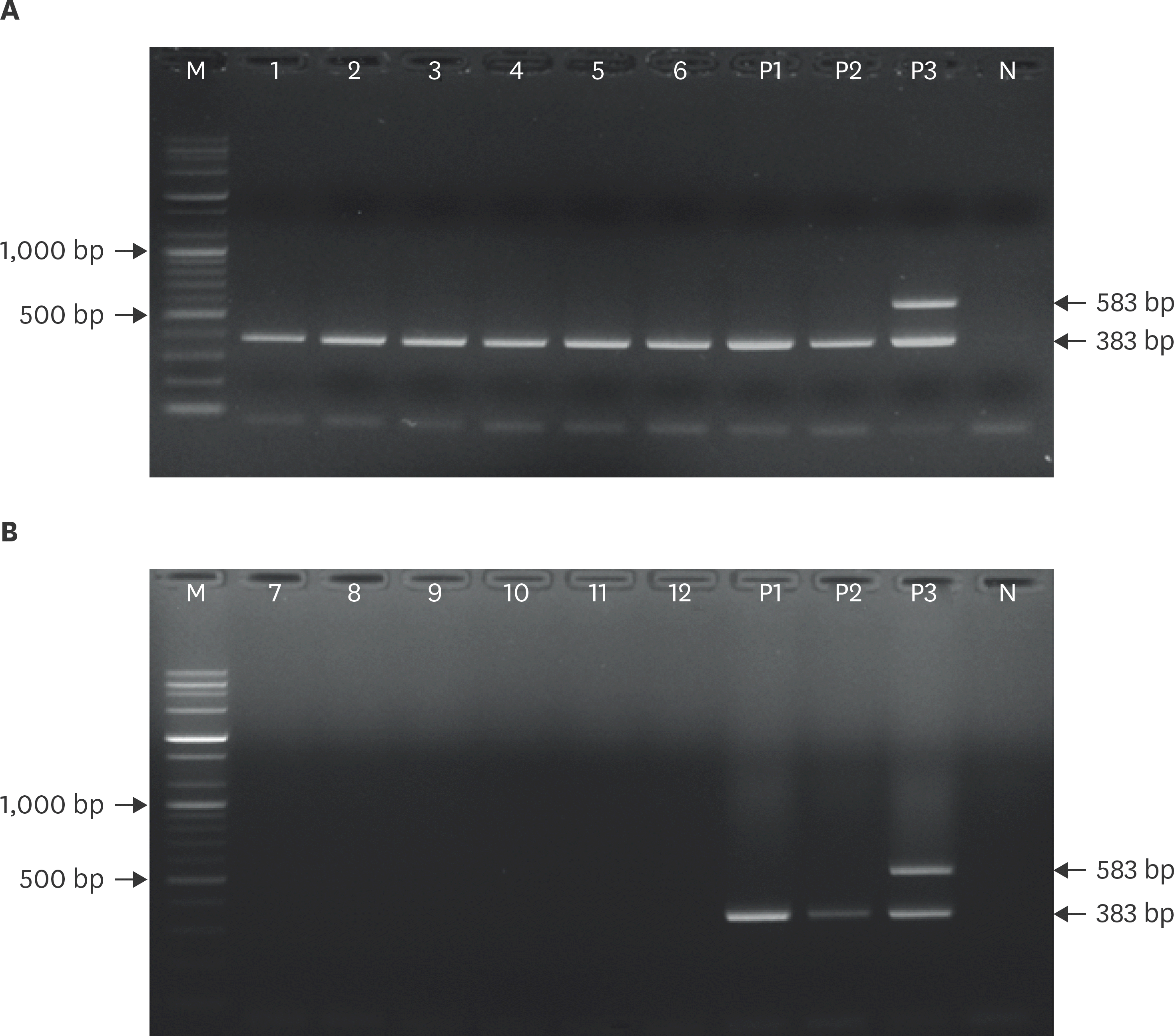 | Fig. 4.Application of multiplex reverse transcription polymerase chain reaction to positive (A) and negative (B) samples. M, 100 bp DNA ladder; lanes 1–6, FAT-positive samples; lane P1, CVS11; lane P2, Evelyn-Rokitnicki-Abelseth strain; lane P3, ERAGS strain; lanes 7–12, FAT-negative samples; lane N, negative control (water only). FAT, fluorescent antibody test. |
Table 1.
Two specific primer sets for differentiation between ERAGS and fixed strains by multiplex reverse transcription polymerase chain reaction
| Designation | Oligonucleotide | Expected size (bp) | Target gene | Remarks |
|---|---|---|---|---|
| RABVcomF | CCCTCCTY* CAGCAACATATGGAG | 383 | G | All RABV |
| RABVcomR | TCACAGTCTGGTCTCACCR* CC | |||
| ERAGS-F | ACCAGACAAGCTTGGTCCCTGG | 583 | G | ERAGS only |
| ERAGS-R | GGA† TGCTCTCTTCCCTCTACTGGA |
Table 2.
Infectivity titer equivalent of multiplex RT-PCR for detection of rabies virus




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download