Abstract
Given that the current Newcastle disease virus (NDV) infection in wild birds poses the threat to poultry, surveillance of Newcastle disease in captive wild birds was carried out in Jilin, China in 2018. Here, an NDV strain obtained from toco toucan was firstly characterized. The results showed that the F gene of the NDV isolate Toucan/China/3/2018 is classified as genotype II in class II. Sequence analysis of the F0 cleavage site was 113 RQGR/L117, which supports the result of the intracerebral pathogenicity index assay indicating classification of the isolate as low-pathogenicity. Experimental infection demonstrated that Toucan/ China/3/2018 can effectively replicate and transmit among chickens. To our knowledge, this is the first report on genetically and pathogenically characterizing NDV strain isolated from toucan, which enriches the epidemiological information of NDV in wild birds.
Go to : 
References
1. Napp S, Alba A, Rocha AI, Sánchez A, Rivas R, Majó N, Perarnau M, Massot C, Miguel ES, Soler M, Busquets N. Six-year surveillance of Newcastle disease virus in wild birds in north-eastern Spain (Catalonia). Avian Pathol. 2017; 46:59–67.

2. Maes P, Amarasinghe GK, Ayllón MA, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chandran K, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Domier LL, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PB, Fouchier RA, Freitas-Astúa J, Ghedin E, Griffiths A, Hewson R, Horie M, Hurwitz JL, Hyndman TH, Jiāng D, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lee B, Leroy EM, Lǐ J, Marzano SL, Mühlberger E, Netesov SV, Nowotny N, Palacios G, Pályi B, Pawęska JT, Payne SL, Rima BK, Rota P, Rubbenstroth D, Simmonds P, Smither SJ, Song Q, Song T, Spann K, Stenglein MD, Stone DM, Takada A, Tesh RB, Tomonaga K, Tordo N, Towner JS, van den Hoogen B, Vasilakis N, Wahl V, Walker PJ, Wang D, Wang LF, Whitfield AE, Williams JV, Yè G, Zerbini FM, Zhang YZ, Kuhn JH. Taxonomy of the order Mononegavirales: second update 2018. Arch Virol. 2019; 164:1233–1244.

3. Dimitrov KM, Abolnik C, Afonso CL, Albina E, Bahl J, Berg M, Briand FX, Brown IH, Choi KS, Chvala I, Diel DG, Durr PA, Ferreira HL, Fusaro A, Gil P, Goujgoulova GV, Grund C, Hicks JT, Joannis TM, Torchetti MK, Kolosov S, Lambrecht B, Lewis NS, Liu H, Liu H, McCullough S, Miller PJ, Monne I, Muller CP, Munir M, Reischak D, Sabra M, Samal SK, Servan de Almeida R, Shittu I, Snoeck CJ, Suarez DL, Van Borm S, Wang Z, Wong FY. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol. 2019; 74:103917.

4. Alexander DJ. Gordon memorial lecture. Newcastle disease. Br Poult Sci. 2001; 42:5–22.
5. Alexander DJ. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 2011; 40:547–558.

6. Pearson GL, McCann MK. The role of indigenous wild, semidomestic, and exotic birds in the epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1972–1973. J Am Vet Med Assoc. 1975; 167:610–614.
7. Newcastle disease (infection with Newcastle disease virus). World Organisation for Animal Health. (ed.).Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 8th ed.pp.p. 964–983. OIE;Paris: 2018.
8. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.

9. Ding Z, Cong YL, Chang S, Wang GM, Wang Z, Zhang QP, Wu H, Sun YZ. Genetic analysis of avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from swine populations in China related to commonly utilized commercial vaccine strains. Virus Genes. 2010; 41:369–376.

Go to : 
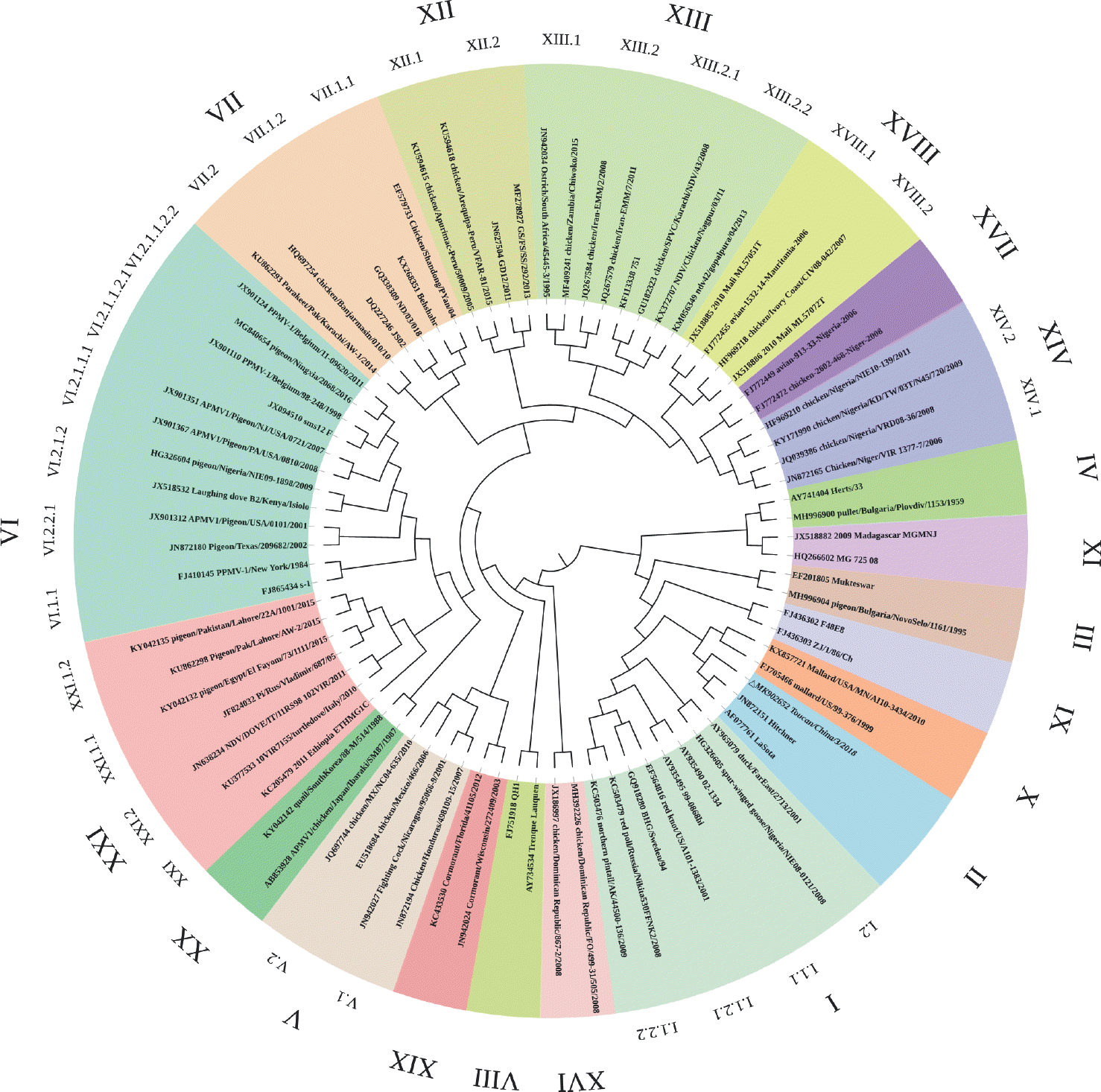 | Fig. 1.Phylogenetic analysis based on the open reading frame (1–1,662 nt) of F genes from 20 genotypes of Newcastle disease virus. The tree was created by the maximum-likelihood method and bootstrapped with 1,000 replicates. Toucan isolate analyzed in the present study is in italics. |
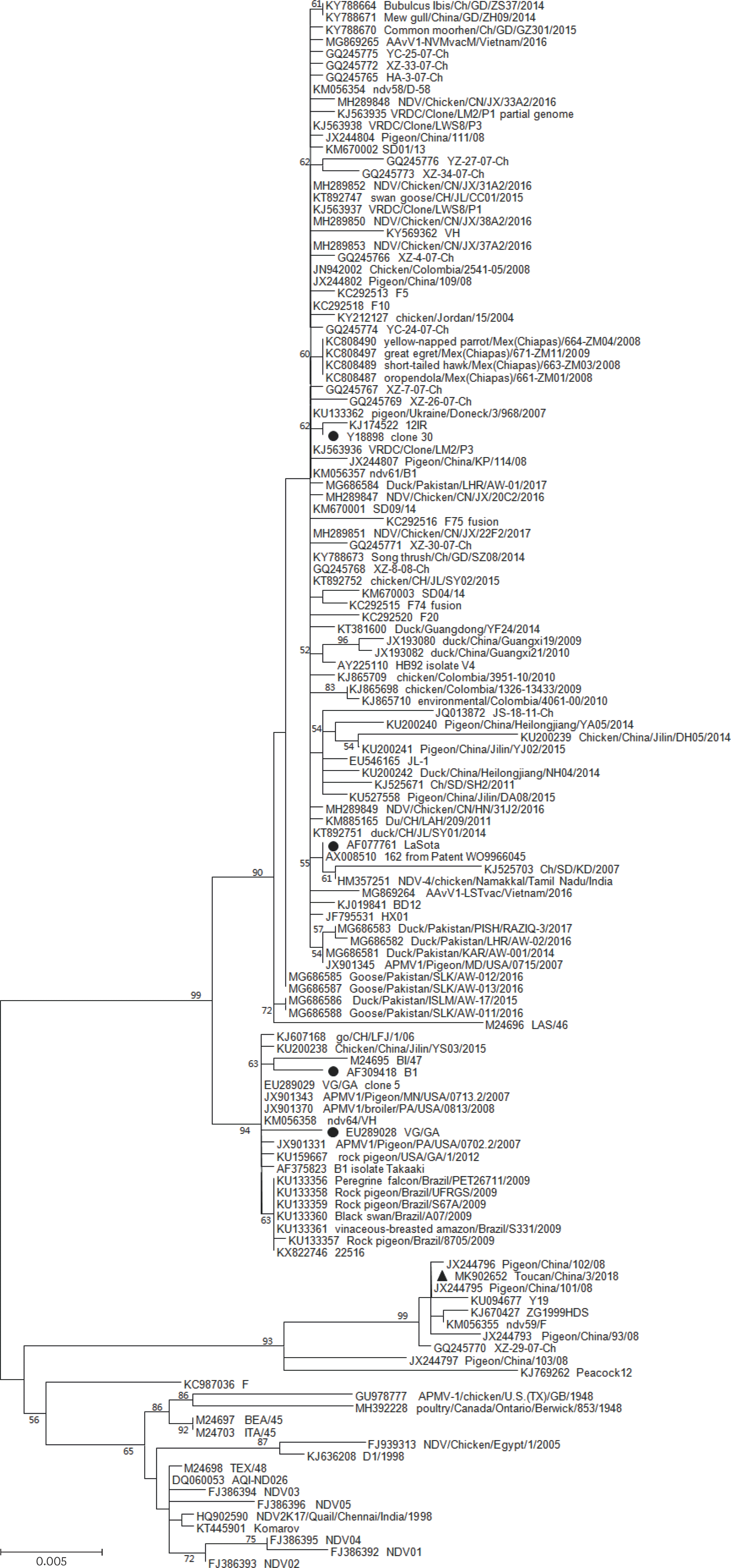 | Fig. 2.Phylogenetic analysis based on the full-length (1–1,662 nt) sequence of F genes from genotype II of Newcastle disease virus available in GenBank. The tree was created by the maximum-likelihood method and bootstrapped with 1,000 replicates. Only bootstrap values above 50 are shown. The vaccine strains are shown as solid black circles. Toucan strain analyzed in the present study is shown as a solid black triangle. |
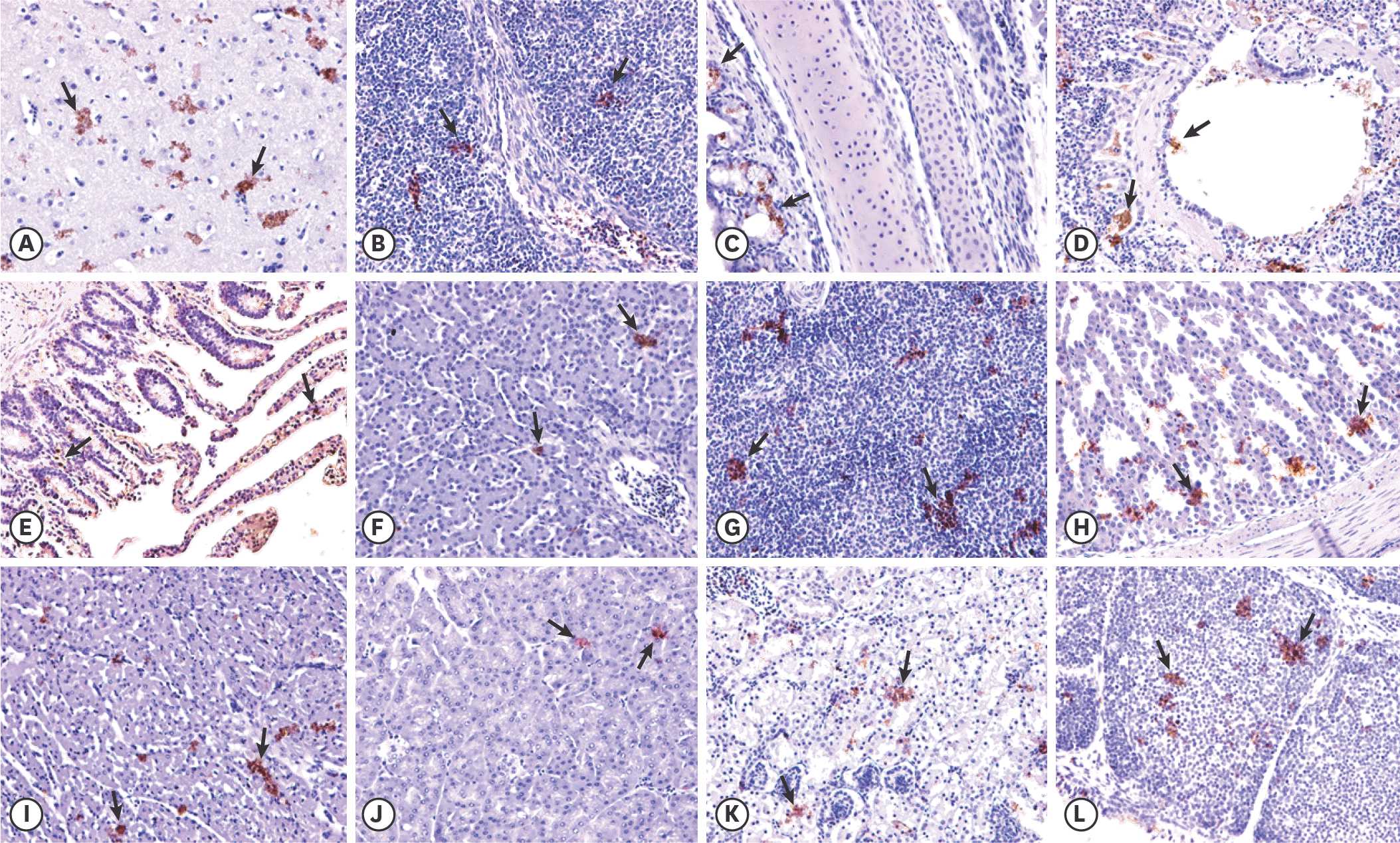 | Fig. 3.Immunohistochemistry analysis of tissue samples from chickens intranasally inoculated with Toucan/China/3/2018 at 7 days post inoculation. Arrows indicate the positive labeling of Newcastle disease virus. (A) Brain; (B) thymus; (C) trachea; (D) lung; (E) heart; (F) liver; (G) spleen; (H) glandular stomach; (I) small intestine; (J) pancreas; (K) kidney; and (L) bursa of Fabricius. Arrows indicate magnification, 20×. |
Table 1.
Primers used for amplifying the Newcastle disease virus strain of Toucan/China/3/2018 in the study
Table 2.
Genomic characteristics of Toucan/China/3/2018
Table 3.
Frequency of NDV isolation and virus titers from chickens challenged and those for cohabitation infection
Table 4.
Comparison of amino acid substitutions in the functional domains of the F protein
| Strains | HRc (471–500 aa) | Major transmembrane domain (501–521 aa) |
|---|---|---|
| 486 | 520 | |
| LaSota | R | I |
| Toucan/China/3/2018 | S | V |
Table 5.
Comparison of amino acid substitutions in the functional domains of the HN protein




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download