Abstract
Objectives
The purpose of this study was to evaluate the association between the distribution of periodontopathic bacteria and oral conditions.
Methods
Stimulated saliva was collected from 162 adults aged 60 years and above. The prevalence and amount of periodontopathic bacteria in the saliva were analyzed using real-time PCR. Pocket depth and clinical attachment loss were examined to evaluate the oral conditions of the subjects. Patients who had at least one tooth surface involved, with a pocket depth ≥4 mm or clinical attachment loss ≥5 mm were classified as having periodontal disease.
Results
The detection rates of most bacteria in the orange and green complexes were more than 90%, while those of P. gingivalis and T. forsythia in the red complex were 58.6% and 61.7%, respectively. The number of bacteria in the red complex positively correlated with each other. There were no significant differences in the number of types of red complex bacteria in the saliva and the distribution of residual number of teeth and periodontal disease (P>0.05). On the other hand, the number of remaining teeth in subjects with higher bacterial density were more than those with lesser bacterial density. In addition, the occurrence of dental disease differed significantly depending on the number of P. gingivalis and T. forsythia.
Go to : 
References
1. Darveau RP. Peiodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010; 8:481–490.
2. Offenbacher S, Barros SP, Beck JD. Rethinking periodontal inflammation. J Periodontol. 2008; 79:1577–1584.

5. Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992; 63:322–331.

6. Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, et al. Oral biofilm architecture on natural teeth. PloS one. 2010; 24(5):e9321.

7. Loomer PM. Microbiological diagnostic testing in the treatment of periodontal diseases. Periodontol 2000. 2004; 34:49–56.

8. Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991; 18:729–739.

9. Tanner AC, Socransky SS, Goodson JM. Microbiota of periodontal pockets losing crestal alveolar bone. J Periodontal Res. 1984; 19:279–291.

10. Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994; 5:78–111.

11. Yucel-Lindberg T, Båge T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev Mol Med. 2013; 15:e7.

12. Genco RJ. Host responses in periodontal diseases: current concepts. J Periodontol. 1992; 63(4 Suppl):S338–355.

13. Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J periodontol. 2003; 74:391–401.

14. Beklen A, Tüter G, Sorsa T, Hanemaaijer R, Virtanen I, Tervahartiala T, et al. Gingival tissue and crevicular fluid co-operation in adult periodontitis. J Dent Res. 2006; 85:59–63.

15. Gersdorf H, Meissner A, Pelz K, Krekeler G, Gobel UB. Identification of Bacteroides forsythus in subgingival plaque from patients with advanced periodontitis. J Clin Microbiol. 1993; 31:941–946.

16. Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000; 38:2362–2365.
17. Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol Immunol. 2001; 45:39–44.

18. Ximénez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra-and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000; 27:722–732.
19. Mayanagi G, Sato T, Shimauchi H, Takahashi N. Detection frequency of periodontitis-associated bacteria by polymerase chain reaction in subgingival and supragingival plaque of periodontitis and healthy subjects. Oral Microbiol Immunol. 2004; 19:379–385.

20. Kulekci G, Leblebicioglu B, Keskin F, Ciftci S, Badur S. Salivary detection of periodontopathic bacteria in periodontally healthy children. Anaerobe. 2008; 14:49–54.

21. Choi HJ, Kim JH, Lee DW, Yang YM, Kim JG. Prevalence of peri-odontopathogens in saliva and plaque of Korean children and adolescents. J Korean Acad Pediatr Dent. 2016; 43:8–16.

22. Kim SM, Yang KH, Choi NK, Kang MS, Oh JS. Quantitative detection of periodontopathic bacteria using real-time PCR. J Korean Acad Pediatr Dent. 2008; 35:494–503.
23. Park SH, Kim SY, Choi SH, Chai JK, Kim CK, Cho KS. The detection of subgingival plaque microflora using 16S rRNA analysis in Korean adult periodontitis. J Korean Acad Periodontol. 1998; 28:691–701.

24. Kim YS, Lee JH, Lee YE. Comparison of quantitative detection of periodontal pathogens before and after scaling by real-time polymerase chain reaction. J Korean Soc Dent Hyg. 2015; 15:1063–1071.

25. Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods. 2004; 59:117–125.

26. Yun JH, Park JE, Kim DI, Lee SI, Choi SH, Cho KS, et al. Identification of putative periodontal pathogens in Korean chronic periodontitis patients. J Korean Acad Periodontol. 2008; 38:143–152.

27. Darby I, Curtis M. Microbiology of periodontal disease in children and young adults. Periodontology. 2000; 26:33–53.

28. Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol 2000. 1999; 20:82–121.

29. Kawada M, Yoshida A, Suzuki N, Nakano Y, Saito T, Oho T, et al. Prevalence of Porphyromonas gingivalis in relation to periodontal status assessed by real-time PCR. Oral Microbiol Immunol. 2004; 19:289–292.

30. Asai Y, Jinno T, Igarashi H, Ohyama Y, Ogawa T. Detection and quantification of oral treponemes in subgingival plaque by realtime PCR. J Clin Microbiol. 2002; 40:3334–3340.

31. Suzuki N, Yoshida A, Saito T, Kawada M, Nakano Y. Quantitative microbiological study of subgingival plaque by real-time PCR shows correlation between levels of Tannerella forsythensis and Fusobacterium spp. J Clin Microbiol. 2004; 42:2255–2257.
Go to : 
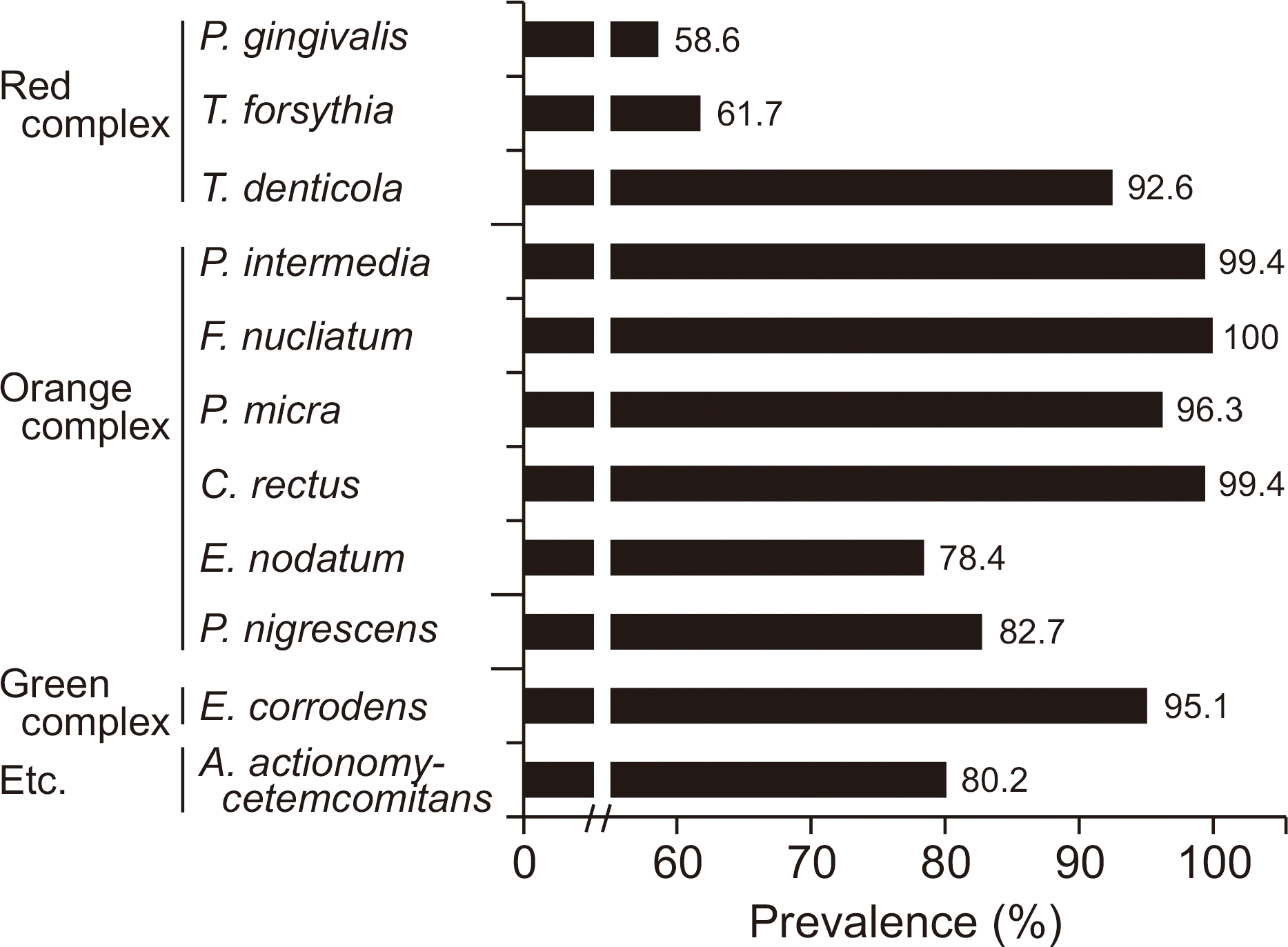 | Fig. 1.Prevalence of periodontopathic bacteria in study participants by multiplex real-time PCR. |
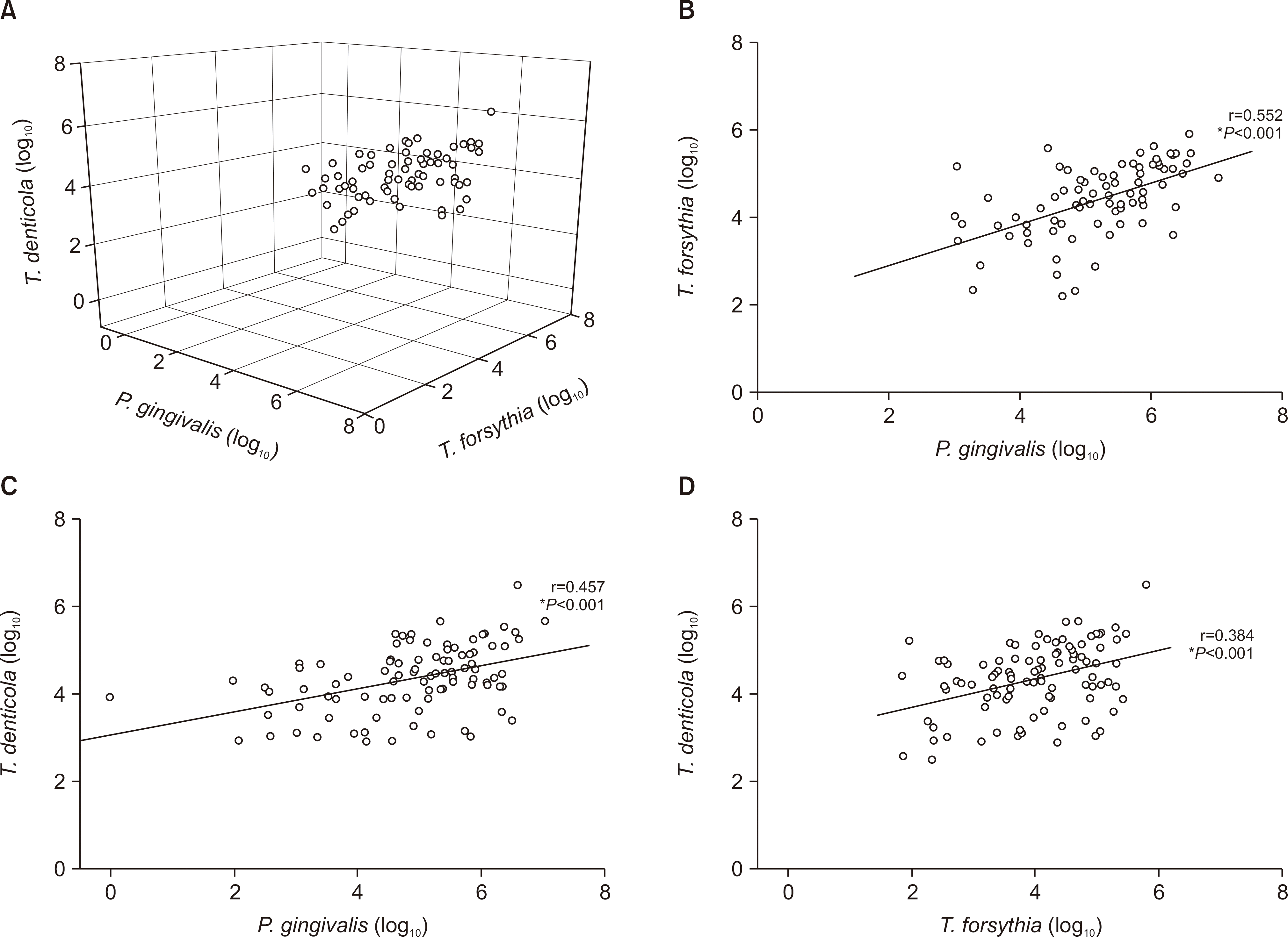 | Fig. 2.Correlation of red complex bacteria. *P-value was analyzed by Pearson's correlation. |
Table 1.
General characteristics of the study participants
Table 2.
Amount of periodontopathic bacteria in study participants by multiplex real-time PCR (Mean±S.D.)
Table 3.
Oral health status according to red complex bacteria species
| The number of strains | N (%) | Number of remaining teeth |
Periodontitis |
|
|---|---|---|---|---|
| No | Yes | |||
| 0 | 7 (4.3) | 14.29±7.59 | 1 (3.2) | 6 (5.3) |
| 1 | 42 (25.9) | 15.21±10.77 | 8 (25.8) | 27 (23.7) |
| 2 | 36 (22.2) | 18.67±10.97 | 8 (25.8) | 23 (20.2) |
| 3 | 77 (47.5) | 19.08±8.47 | 14 (45.2) | 58 (50.9) |
| Total | 162 (100) | 17.78±9.74 | 31 (100) | 114 (100) |
| P-value | 0.139* | 0.855† | ||
Table 4.
Oral health status according to the number of red complex bacteria
| Division | The number of bacteria | N (%) | The number of remaining teeth |
Periodontitis‡ |
|
|---|---|---|---|---|---|
| No | Yes | ||||
| P. gingivalis | |||||
| <3.8×105 | 133 (82.1) | 16.91±10.13 | 29 (93.5) | 88 (77.2) | |
| ≥3.8×105 | 29 (17.9) | 21.76±6.50 | 2 (6.5) | 26 (22.8) | |
| P-value | 0.002* | 0.041† | |||
| T. forsythia | |||||
| <4.4×104 | 126 (77.8) | 16.67±10.18 | 28 (19.3) | 82 (71.9) | |
| ≥4.4×104 | 36 (22.2) | 21.64±6.84 | 3 (9.7) | 32 (28.1) | |
| P-value | 0.001* | 0.034† | |||
| T. denticola | |||||
| <7.1×104 | 134 (82.7) | 17.10±10.07 | 27 (87.1) | 91 (79.8) | |
| ≥7.1×104 | 28 (17.3) | 21.00±7.32 | 4 (12.9) | 23 (20.2) | |
| P-value | 0.021* | 0.356† | |||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download