INTRODUCTION
METHODS
Data source
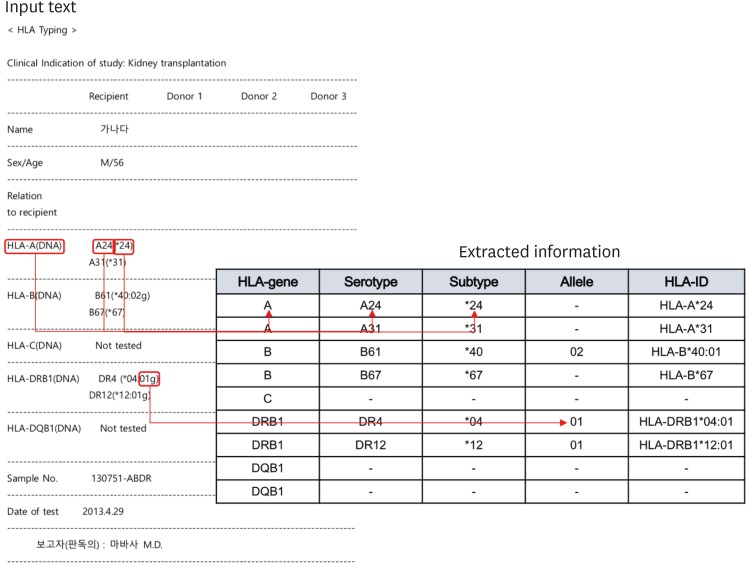
 | Fig. 1Representative example of raw data in an HLA report. A real example of an HLA typing report with de-identified patient names is shown. An HLA typing result is represented in one cell in an Excel file. This example includes HLA typing results for three patients because most HLA test subjects were recipients of a transplantation procedure, and the physicians wanted to compare all HLA tests of candidate donors with those of a given recipient on the same page on the electronic medical record. In this cell, the HLA test results were arranged in a tabular form using the space bar and a carriage return but were not structured as actual tables with distinct rows and columns. To improve accuracy, we only focused on reports with the HLA typing results of one patient.HLA = human leukocyte antigen.
|
Data extraction method
Performance evaluation
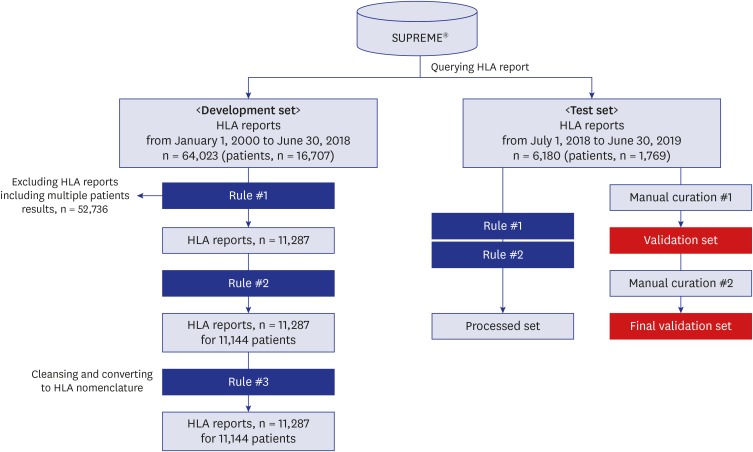 | Fig. 2HLA typing and clinical characteristics extraction pipeline. Rule #1 was designed to exclude HLA reports with typing results for multiple patients. After excluding these reports, we applied Rule #2 to extract clinical variables such as name, sex, and indications of HLA typing. Rule #1 and Rule #2 are extraction rules, and Rule #3 is cleaning rule. Rule #3 was designed to clean the HLA typing results and transform the results to a standard nomenclature. To evaluate the accuracy of the two extraction rules, we applied Rule #1 and Rule #2 to the testing set. The rule-based extraction results of the testing set were then compared with the manually curated results of the testing test, as the validation set. The manual curation process was done sequentially by two different investigators.HLA = human leukocyte antigen.
|
Cleaning and converting the HLA typing to nomenclature
Ethics statement
RESULTS
Summary of HLA typing data
HLA data extraction rules
Table 1
Typical Python expressions used to extract the HLA genotype status and clinical variables for each patient
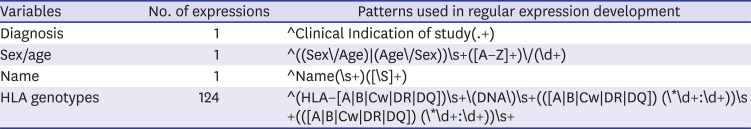
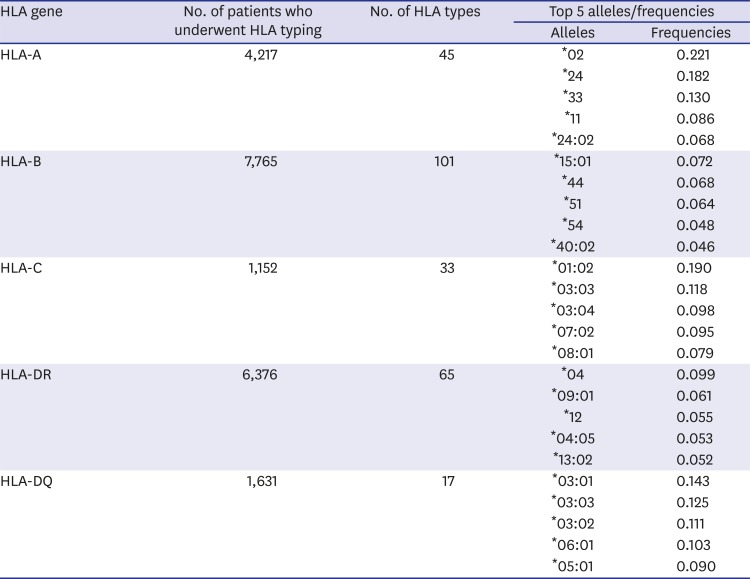
Table 2
HLA genotype frequencies in the test set extracted by Rule #1 and Rule #2
Validation of NLP accuracy
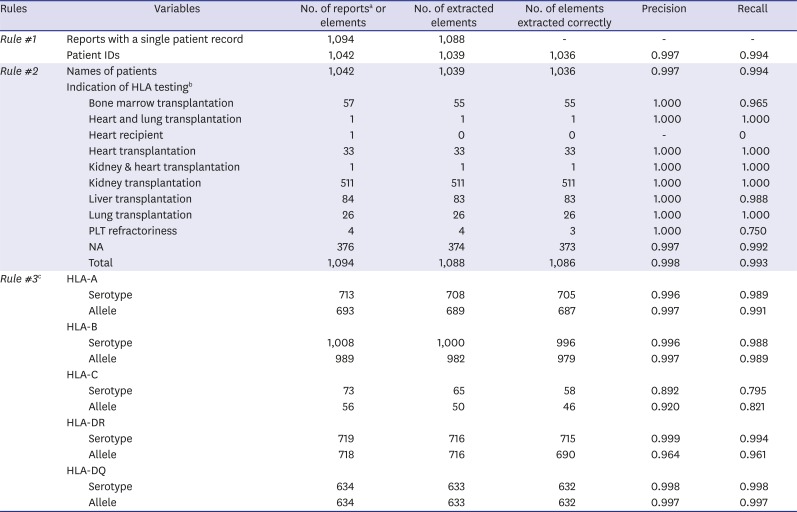
Table 3
Recall and precision for clinical variables and serotype/alleles of HLA genes




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download