INTRODUCTION
MATERIALS AND METHODS
Materials
Cell culture and treatment
Cell proliferation and viability assay
Nitric oxide production assay
Western blot assay
Total RNA isolation Assay
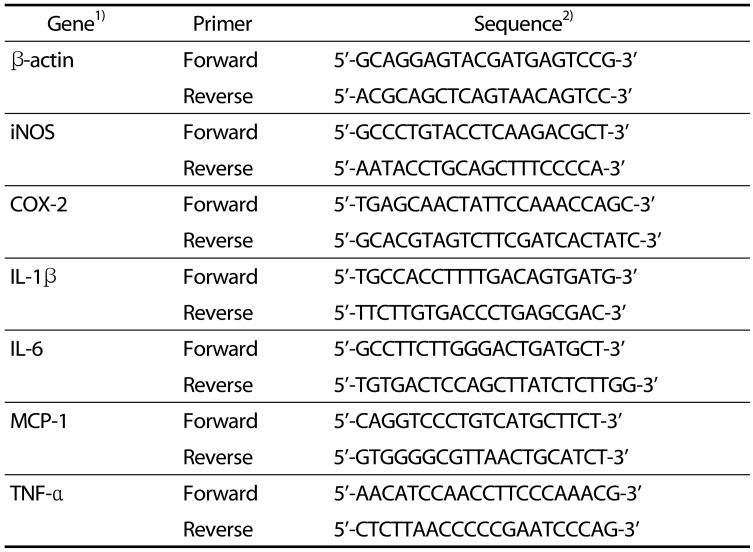
Table 1
Primer sequences used in real-time PCR analysis.
Statistical analysis
RESULTS
Effect of saccharin on adipocyte proliferation
 | Fig. 1Effect of saccharin on cell proliferation in 3T3-L1 adipocytes.(A) 3T3-L1 cells were plated at a density of 1.0 × 104 cells/mL in a 24 well plate. After 4 days of incubation, the media were replaced with differentiation media to induce differentiation into adipocytes. Obtained adipocytes were then treated with saccharin (0, 50, 100, and 200 µg/mL) for 0, 24, 48, 96, 144 hours, cell viability was estimated by MTT assay. (B) After adipocytes were treated with the saccharin (0, 50, 100, and 200 µg/mL) for 24 h and LPS (1 µm/mL) for a further 18 h, cell viability was estimated by MTT assay. Each bar presents the mean ± SE (n = 3) and different letters indicate significant differences among treatments at P < 0.05 (Duncan's multiple range test).
|
Assay for NO production mRNA expression of iNOS
 | Fig. 2Effect of saccharin on NO production and mRNA expression of iNOS in LPS-stimulated 3T3-L1 adipocytes.3T3-L1 cells were plated at a density of 1.0 × 104 cells/mL in a 24-well plate. After 4 days of incubation, the media were replaced with differentiation media to induce differentiation into adipocytes. After adipocytes were treated with the saccharin (0, 50, 100, and 200 µg/mL) for 24 h and LPS (1 µm/mL) for a further 18 h. (A) Production of nitric oxide (NO) was determined by using Griess reagent. (B) Total mRNA expression of inducible nitric oxide synthase (iNOS) was determined by performing real-time PCR. Each bar presents the mean ± SE (n = 3) and different letters indicate significant differences among treatments at P < 0.05 (Duncan's multiple range test).
|
The protein expression levels of NF-κB and IκB
 | Fig. 3Effect of saccharin on protein expressions of NF-κB and IκB in LPS-stimulated 3T3-L1 adipocytes.3T3-L1 cells were plated at a density of 1.0 × 104 cells/mL in a 24-well plate. After 4 days of incubation, the media were replaced with differentiation media to induce differentiation into adipocytes. After adipocytes were treated with the saccharin (0, 50, 100, and 200 µg/mL) for 24 h and LPS (1 µm/mL) for a further 18 h. (A) The protein expression of nuclear factor kappa B (NF-κB) was measured by western blot analysis. (B) The protein expression of inhibitor κB (IκB) was measured by western blot analysis. Each bar presents the mean ± SE (n = 3) and different letters indicate significant differences among treatments at P < 0.05 (Duncan's multiple range test).
|
mRNA expressions of inducible COX-2
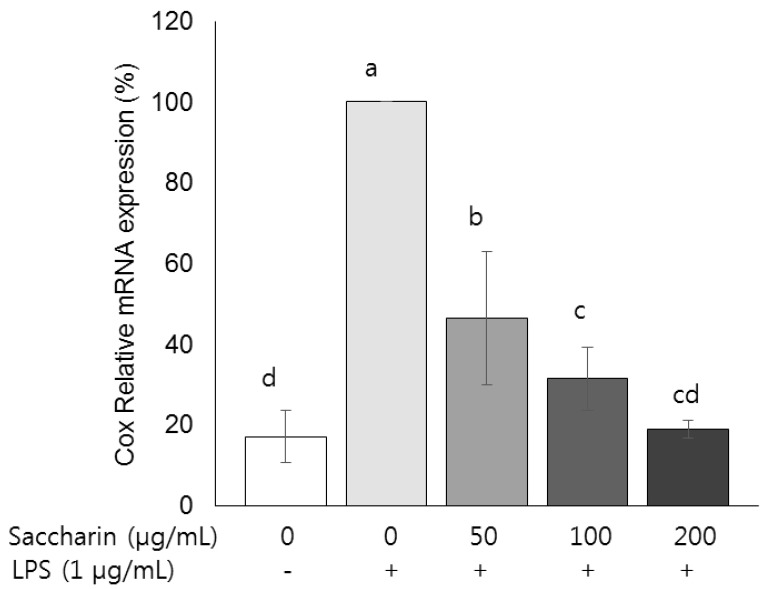 | Fig. 4Effect of saccharin on the mRNA expression of COX-2 in LPS-stimulated 3T3-L1 adipocytes.3T3-L1 cells were plated at a density of 1.0 × 104 cells/mL in a 24-well plate. After 4 days of incubation, the media were replaced with differentiation media to induce differentiation into adipocytes. After adipocytes were treated with the saccharin (0, 50, 100, and 200 µg/mL) for 24 h and LPS (1 µm/mL) for a further 18 h. The mRNA expression of cyclooxygenase-2 (COX-2) was determined by performing real-time PCR. Each bar presents the mean ± SE (n = 3) and different letters indicate significant differences among treatments at P < 0.05 (Duncan's multiple range test).
|
mRNA expression of inflammatory cytokines IL-1β, IL-6, MCP-1, and TNF-α
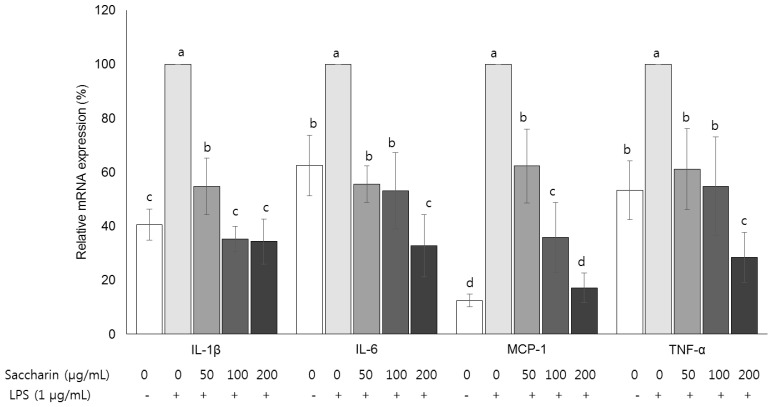 | Fig. 5Effect of saccharin on the mRNA expression of cytokines in LPS-stimulated 3T3-L1 adipocytes.3T3-L1 cells were plated at a density of 1.0 × 104 cells/mL in a 24-well plate. After 4 days of incubation, the media were replaced with differentiation media to induce differentiation into adipocytes. After adipocytes were treated with the saccharin (0, 50, 100, and 200 µg/mL) for 24 h and LPS (1 µm/mL) for a further 18 h. The mRNA expressions of interleukin 1β (IL-1β), interleukin 6 (IL-6), monocyte chemoattractant protein-1 (MCP-1), and tumor necrosis factor-α (TNF-α) were determined by performing real-time PCR. Each bar presents the mean ± SE (n = 3) and different letters indicate significant differences among treatments at P < 0.05 (Duncan's multiple range test).
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download