Linezolid is an oxazolidinone that binds 23S rRNA and inhibits protein synthesis by preventing 70S ribosomal unit formation [
1]. Since its introduction in 2000, linezolid has been the last-resort antimicrobial for vancomycin-resistant enterococci and methicillin-resistant
Staphylococcus aureus [
1]. Linezolid-resistant
Enterococcus faecium was first observed in patients infected with vancomycin-resistant
E. faecium who were treated with linezolid for a prolonged period. The resistance was due to mutations in domain V of the 23S rRNA of
E. faecium [
2]. While this mechanism of linezolid resistance in
E. faecium and
S. aureus is predominant in Korea, resistance due to variants in ribosomal proteins L3, L4, and L22 have also been reported [
3]. Acquisition of the linezolid resistance gene
cfr or, more recently,
optrA is more threatening because these genes are plasmid encoded and, thus, transmissible [
4]. Human isolates of
optrA-positive linezolid-nonsusceptible
Enterococcus faecalis (LNSEF) were reported first in 2016 in China [
4] and later in the USA, Sweden, France, Thailand, Taiwan, Malaysia, and Korea [
35]. However,
optrA has been detected among archival clinical isolates traceable to 2012 in Korea [
3]. In Asan Medical Center, a 2,715-bed, university-affiliated tertiary care hospital in Seoul, Korea, LNSEF was first detected in November 2017. There is a lack of data on LNSEF in Korea. In this study, we investigated the resistance mechanisms and epidemiology of LNSEF isolates.
Clinical isolates of Enterococcus species collected at the Asan Medical Center were subjected to species identification and antimicrobial susceptibility testing using the MicroScan PBC 28 panel (Beckman Coulter, Brea, CA). Linezolid susceptibility was confirmed by the E-test (bioMérieux SA, Marcy l'Etoile, France). LNSEF isolates were collected for 20 months, from November 2017 to June 2019.
The molecular mechanism for linezolid resistance was investigated by 23S rRNA gene sequencing and
cfr or
optrA-specific PCR. For molecular epidemiological analysis, seven housekeeping genes (
gdh,
gyd,
pstS,
gki,
aroE,
xpt, and
yiqL) were sequenced using PCR, and sequence types (STs) were determined by comparison with an open-access database (PubMLST,
https://pubmlst.org/efaecalis/). The primers were adopted from a previous report [
6]. Six LNSEF strains isolated in the first year until October 2018 were subjected to pulsed-field gel electrophoresis (PFGE) typing with SmaI restriction (Takara, Tokyo, Japan) using the CHEF Mapper system (Bio-Rad, Hercules, CA, USA). Clonality based on PFGE analysis was interpreted according to a previous report [
7].
Demographic findings and clinical history, including infection, operation, antimicrobial treatment, and microbiological data, were obtained from electronic medical records of the patients. This study was approved by the Institutional Review Board of Asan Medical Center (S2019-1169-0001).
In total, 4,318
E. faecalis isolates were obtained from 2,639 patients. Ten isolates (0.23%) were found to be linezolid-nonsusceptible and included five linezolid-intermediate (0.115%) and five linezolid-resistant (0.115%) isolates. Minimum inhibitory concentrations of linezolid as determined by E-tests were 6–12 µg/mL, which were resistant. The isolates were obtained from urine (N=6), Jackson Pratt drainage (N=2), and open pus (N=2) samples. The antimicrobial susceptibility and resistance genes of the LNSEF isolates are listed in
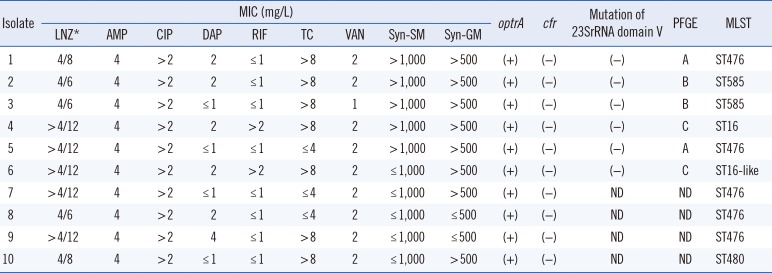
Table 1. All LNSEF isolates were susceptible to penicillin, ampicillin, daptomycin, nitrofurantoin, teicoplanin, and vancomycin, but resistant to fluoroquinolone and quinupristin/dalfopristin.
Table 1
Antibiogram, resistance genes, and molecular epidemiology of LNSEF
|
Isolate |
MIC (mg/L) |
optrA |
cfr |
Mutation of 23SrRNA domain V |
PFGE |
MLST |
|
LNZ*
|
AMP |
CIP |
DAP |
RIF |
TC |
VAN |
Syn-SM |
Syn-GM |
|
1 |
4/8 |
4 |
>2 |
2 |
≤1 |
>8 |
2 |
>1,000 |
>500 |
(+) |
(−) |
(−) |
A |
ST476 |
|
2 |
4/6 |
4 |
>2 |
2 |
≤1 |
>8 |
2 |
>1,000 |
>500 |
(+) |
(−) |
(−) |
B |
ST585 |
|
3 |
4/6 |
4 |
>2 |
≤1 |
≤1 |
>8 |
1 |
>1,000 |
>500 |
(+) |
(−) |
(−) |
B |
ST585 |
|
4 |
>4/12 |
4 |
>2 |
2 |
>2 |
>8 |
2 |
>1,000 |
>500 |
(+) |
(−) |
(−) |
C |
ST16 |
|
5 |
>4/12 |
4 |
>2 |
≤1 |
≤1 |
≤4 |
2 |
>1,000 |
>500 |
(+) |
(−) |
(−) |
A |
ST476 |
|
6 |
>4/12 |
4 |
>2 |
2 |
>2 |
>8 |
2 |
≤1,000 |
>500 |
(+) |
(−) |
(−) |
C |
ST16-like |
|
7 |
>4/12 |
4 |
>2 |
≤1 |
≤1 |
≤4 |
2 |
≤1,000 |
>500 |
(+) |
(−) |
ND |
ND |
ST476 |
|
8 |
4/6 |
4 |
>2 |
2 |
≤1 |
≤4 |
2 |
≤1,000 |
≤500 |
(+) |
(−) |
ND |
ND |
ST476 |
|
9 |
>4/12 |
4 |
>2 |
4 |
≤1 |
>8 |
2 |
≤1,000 |
≤500 |
(+) |
(−) |
ND |
ND |
ST476 |
|
10 |
4/8 |
4 |
>2 |
≤1 |
≤1 |
>8 |
2 |
≤1,000 |
>500 |
(+) |
(−) |
ND |
ND |
ST480 |

All 10 LNSEF isolates were positive for
optrA, but none were positive for
cfr. The domain V of the 23S rRNA genes of all six isolates in the first year were wild-type. Multilocus sequence typing (MLST) revealed that five isolates were ST476, two ST585, one ST16, one ST16-like, and one ST480. The pulsotypes of the six isolates in the first year were clustered into three types (
Fig. 1). The ST16-like isolate harbored a thymine at the 27th position of
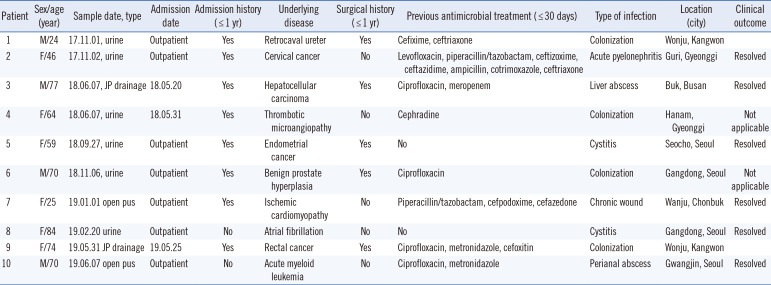
yiqL, while ST16 had cytosine. Clinical and epidemiological features of the 10 patients with LNSEF are presented in
Table 2. Seven of these LNSEF isolates were obtained from urine samples and open pus samples in an outpatient setting, whereas the remaining three were hospital-acquired. In one patient, linezolid-susceptible enterococci were recovered from a urine sample one month before LNSEF isolation. None of the patients carrying LNSEF had been treated with linezolid. Eight patients had a history of hospitalization. Two patients had an overlap in admission period, but they had been treated in different wards, ruling out contact transmission. The five ST476 isolates were from patients who lived in three different cities.
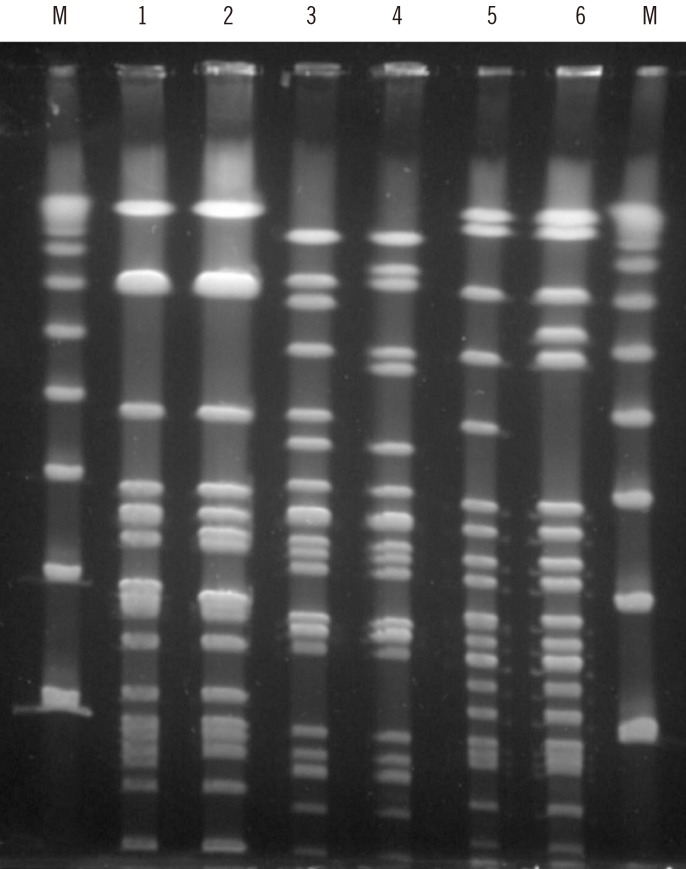 | Fig. 1Pulsed-field gel electrophoresis (PFGE) analysis of linezolid-nonsusceptible Enterococcus faecalis isolates after SmaI-digestion. M denotes Lambda Ladder PFG Marker (New England Biolabs Inc., N0341S, Beverly, MA). Each of lanes from 1 to 6 denoted the pulsotype of isolate number 1, 5, 2, 3, 4, and 6 in order.
|
Table 2
Clinical and epidemiological features of 10 patients from whom LNSEF was isolated
|
Patient |
Sex/age (year) |
Sample date, type |
Admission date |
Admission history (≤1 yr) |
Underlying disease |
Surgical history (≤1 yr) |
Previous antimicrobial treatment (≤30 days) |
Type of infection |
Location (city) |
Clinical outcome |
|
1 |
M/24 |
17.11.01, urine |
Outpatient |
Yes |
Retrocaval ureter |
Yes |
Cefixime, ceftriaxone |
Colonization |
Wonju, Kangwon |
|
|
2 |
F/46 |
17.11.02, urine |
Outpatient |
Yes |
Cervical cancer |
No |
Levofloxacin, piperacillin/tazobactam, ceftizoxime, ceftazidime, ampicillin, cotrimoxazole, ceftriaxone |
Acute pyelonephritis |
Guri, Gyeonggi |
Resolved |
|
3 |
M/77 |
18.06.07, JP drainage |
18.05.20 |
Yes |
Hepatocellular carcinoma |
Yes |
Ciprofloxacin, meropenem |
Liver abscess |
Buk, Busan |
Resolved |
|
4 |
F/64 |
18.06.07, urine |
18.05.31 |
Yes |
Thrombotic microangiopathy |
No |
Cephradine |
Colonization |
Hanam, Gyeonggi |
Not applicable |
|
5 |
F/59 |
18.09.27, urine |
Outpatient |
Yes |
Endometrial cancer |
Yes |
No |
Cystitis |
Seocho, Seoul |
Resolved |
|
6 |
M/70 |
18.11.06, urine |
Outpatient |
Yes |
Benign prostate hyperplasia |
Yes |
Ciprofloxacin |
Colonization |
Gangdong, Seoul |
Not applicable |
|
7 |
F/25 |
19.01.01 open pus |
Outpatient |
Yes |
Ischemic cardiomyopathy |
No |
Piperacillin/tazobactam, cefpodoxime, cefazedone |
Chronic wound |
Wanju, Chonbuk |
Resolved |
|
8 |
F/84 |
19.02.20 urine |
Outpatient |
No |
Atrial fibrillation |
No |
No |
Cystitis |
Gangdong, Seoul |
Resolved |
|
9 |
F/74 |
19.05.31 JP drainage |
19.05.25 |
Yes |
Rectal cancer |
Yes |
Ciprofloxacin, metronidazole, cefoxitin |
Colonization |
Wonju, Kangwon |
|
|
10 |
M/70 |
19.06.07 open pus |
Outpatient |
No |
Acute myeloid leukemia |
No |
Ciprofloxacin, metronidazole |
Perianal abscess |
Gwangjin, Seoul |
Resolved |

In this study, the prevalence of
optrA-positive LNSEF of 0.23% was comparable to those of a linezolid resistance surveillance program (2011 to 2015) in the United States, in which 0.3%–0.7% of enterococci were linezolid-nonsusceptible [
8]. In line with this finding, a recent study in a tertiary care hospital in Korea revealed that 1.65% of enterococci were nonsusceptible to linezolid, and 20.8% of LNSEF isolates carried
optrA, accounting for approximately 0.3% of the total
E. faecalis isolates [
3]. This prevalence of
optrA-mediated linezolid resistance is similar to that observed in the present study. We identified
optrA as the primary mechanism of linezolid resistance in
E. faecalis, whereas during the same period, 23S rRNA mutations was identified as the primary mechanism underlying linezolid resistance in
E. faecium [
9]. Considering the ability of bacteria to transfer
optrA via plasmids and the marked increase in
optrA-positive
E. faecalis cases in China [
4], the spread of
optrA-mediated linezolid resistance could be an imminent threat in Korea.
In this study, all LNSEF isolates except one were multidrug-resistant. The susceptibility rates of LNSEF isolates to ciprofloxacin, high-level gentamicin, and high-level streptomycin were 0.0%, 20.0%, and 50.0%, respectively, whereas the average susceptibility rates of the total E. faecalis isolates to these antimicrobials were 59.9%, 51.4%, and 86.1%, respectively (unpublished data). Except two, all patients underwent antimicrobial treatment within 30 days before LNSEF isolation. Therefore, most cases were healthcare-associated, although they were isolated from an outpatient setting. Antibiotic pressure seemed to trigger the spread of LNSEF, but none of the patients had a history of linezolid treatment. Therefore, linezolid resistance is likely to spread via clonal expansion or horizontal transfer of resistance genes in a community setting, but in a healthcare-associated manner. Increased spread of LNSEF would complicate the antimicrobial treatment of urogenital and wound infections.
ST476 was the most prevalent strain in this study. Previously
optrA-positive ST476 strains were reported only in China, where it has been frequently found in humans and pigs [
101112]. The epidemiological interpretation of these findings needs further investigation. Among other STs, ST16 is an
optrA-carrying clone with a worldwide distribution [
12]. ST585 strains harboring
optrA have been reported in the USA, China, and Germany [
51013]. ST480 strains harboring
optrA have been reported in China, France, Germany, and Belgium [
12131415]. Studies in China have revealed that
optrA is more prevalent in animal enterococcal isolates (15.9%) than human isolates (2.0%) [
10] and that 3.5% of healthy individuals carry
optrA-carrying
E. faecalis, regardless of age [
16]. The
optrA gene has been found from the enterococcal isolates of food animals since 2008 in Korea, and
optrA-positive LNSEF from food animals belong to several STs, of which only ST16 was detected in the present study [
1718]. Therefore, ST16 may be responsible for the spread of
optrA-positive LNSEF in both food animals and humans. All STs in this study have been previously detected in China. Geographical vicinity along with substantial food exchange and extensive human traveling may explain the epidemiological linkage of
optrA-positive
E. faecalis between these two countries.
In this study, none of the patients with LNSEF were epidemiologically related to one another. Most LNSEF infections were of the community-onset and healthcare-associated type. This may herald an era of linezolid resistance by plasmid-mediated resistance gene acquisition. A Chinese hospital reported that as high as 3.93% of
E. faecalis isolates were linezolid-resistant, and all those isolates were positive for
optrA,
cfr, or both [
11]. Therefore, the rapid spread of LNSEF via clonal expansion or horizontal transfer of resistance genes is a possible scenario once it is introduced [
3419].
There were some limitations in this study. First, the number of LNSEF isolates studied was relatively small, because linezolid resistance in E. faecalis is still rare. Second, this was a single-center study; further multi-center studies are needed to confirm the prevalence and epidemiology of optrA-harboring E. faecalis. In conclusion, the emergence of optrA-mediated LNSEF in a community setting is alarming. Therefore, adequate infection control measures and surveillance of linezolid resistance are necessary in Korea.







 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download