1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.


2. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983; 15:10–17.


3. Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014; 15:e268–78.

4. Abu-Rustum NR, Zhou Q, Gomez JD, Alektiar KM, Hensley ML, Soslow RA, et al. A nomogram for predicting overall survival of women with endometrial cancer following primary therapy: toward improving individualized cancer care. Gynecol Oncol. 2010; 116:399–403.


5. Salvesen HB, Haldorsen IS, Trovik J. Markers for individualised therapy in endometrial carcinoma. Lancet Oncol. 2012; 13:e353–61.

6. Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013; 497:67–73.

7. Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol. 2012; 28:523–553.


8. Pyke C, Rømer J, Kallunki P, Lund LR, Ralfkiaer E, Danø K, et al. The gamma 2 chain of kalinin/laminin 5 is preferentially expressed in invading malignant cells in human cancers. Am J Pathol. 1994; 145:782–791.


9. Pyke C, Salo S, Ralfkiaer E, Rømer J, Danø K, Tryggvason K. Laminin-5 is a marker of invading cancer cells in some human carcinomas and is coexpressed with the receptor for urokinase plasminogen activator in budding cancer cells in colon adenocarcinomas. Cancer Res. 1995; 55:4132–4139.

10. Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007; 7:370–380.

12. Liétard J, Musso O, Théret N, L'Helgoualc'h A, Campion JP, Yamada Y, et al. Sp1-mediated transactivation of LamC1 promoter and coordinated expression of laminin-gamma1 and Sp1 in human hepatocellular carcinomas. Am J Pathol. 1997; 151:1663–1672.


13. Zhang Y, Xi S, Chen J, Zhou D, Gao H, Zhou Z, et al. Overexpression of LAMC1 predicts poor prognosis and enhances tumor cell invasion and migration in hepatocellular carcinoma. J Cancer. 2017; 8:2992–3000.

14. Ke HL, Ke RH, Li B, Wang XH, Wang YN, Wang XQ. Association between laminin γ1 expression and meningioma grade, recurrence, and progression-free survival. Acta Neurochir (Wien). 2013; 155:165–171.


15. Hirai Y, Kawaguchi T, Hasumi K, Kitagawa T, Noda T. Establishment and characterization of human cell lines from a serous papillary adenocarcinoma of the endometrium. Gynecol Oncol. 1994; 54:184–195.

16. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47.

19. Kuramoto H, Nishida M, Morisawa T, Hamano M, Hata H, Kato Y, et al. Establishment and characterization of human endometrial cancer cell lines. Ann N Y Acad Sci. 1991; 622:402–421.


20. Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017; 357:eaan2507.

22. Wang L, Guo J, Wang Q, Zhou J, Xu C, Teng R, et al. LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. J Cancer Res Clin Oncol. 2014; 140:1997–2008.


23. Orlandi R, Cattaneo M, Troglio F, Casalini P, Ronchini C, Ménard S, et al. SEL1L expression decreases breast tumor cell aggressiveness in vivo and in vitro. Cancer Res. 2002; 62:567–574.

24. Cattaneo M, Orlandini S, Beghelli S, Moore PS, Sorio C, Bonora A, et al. SEL1L expression in pancreatic adenocarcinoma parallels SMAD4 expression and delays tumor growth in vitro and in vivo. Oncogene. 2003; 22:6359–6368.


25. Liu Q, Chen J, Mai B, Amos C, Killary AM, Sen S, et al. A single-nucleotide polymorphism in tumor suppressor gene SEL1L as a predictive and prognostic marker for pancreatic ductal adenocarcinoma in Caucasians. Mol Carcinog. 2012; 51:433–438.


26. Liu Q, Chen J, Wang J, Amos C, Killary AM, Sen S, et al. Putative tumor suppressor gene SEL1L was downregulated by aberrantly upregulated hsa-miR-155 in human pancreatic ductal adenocarcinoma. Mol Carcinog. 2014; 53:711–721.


27. Šedová L, Školníková E, Hodúlová M, Včelák J, Šeda O, Bendlová B. Expression profiling of Nme7 interactome in experimental models of metabolic syndrome. Physiol Res. 2018; 67:S543–50.

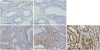
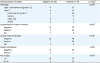







 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download