Abstract
Background
Platelet-rich plasma (PRP) is a plasma component of autologous blood containing a high concentration of platelets. PRP is used to promote healing of damaged tissues. However, there are not many studies on the composition and expression patterns of active proteins in PRP. The purpose of this study was to identify unknown factors that contribute to tissue healing by proteomic analysis of proteins in PRP.
Methods
Three men in their 30s with no basal disease participated in this study. All identified proteins were classified for tissue healing-related functions on the basis of the gene ontology analysis of adhesion molecule with Ig-like domain 2 (AmiGO2). PRP was prepared by using the ACP kit and GPS III kit.
Results
We identified a total of 125 proteins related to wound healing, along with three proteins for angiogenesis involved in wound healing, two proteins for fibroblast migration, four proteins for collagen biosynthesis process, two proteins for glycosaminoglycan biosynthesis process, and 13 proteins for glycosaminoglycan binding. So, in addition to the growth factors that have been already known to be involved in tissue healing, 25 new proteins were identified.
Platelet-rich plasma (PRP) was first used as a component for the transfusion of allogenic blood in open-heart surgery in 1987.1) In the beginning, PRP was used for the preprosthetic procedure and implant placement in oral surgery and as an adjuvant to aid in bone regeneration in maxillofacial surgery. Thereafter, application of PRP has been reported in various fields of medicine including dermatology, plastic surgery, otorhinolaryngology, urology, and neurosurgery.2) In orthopedics and sports medicine, PRP is used to promote healing of damaged tissues through a direct injection of PRP into injury sites. Further studies investigated the use of PRP in acute ligament injuries such as medial collateral ligament injury of the knee and rupture of the lateral ligaments of the ankle and rotator cuff tears, as well as in chronic tendon injury including lateral epicondylitis, patellar tendinitis, plantar fasciitis, Achilles tendinitis, and acute muscle ruptures.345)
PRP is a plasma component of autologous blood with large numbers of platelets. It generally has 3- to 5-fold higher platelet concentrations than whole blood.3) Platelets contain α-granules, dense granules, and lysosomal granules, of which α-granules include a large quantity of bioactive proteins (growth factors and cytokines) and are known to play important roles in hemostasis and tissue healing.67) Thus, the fact that PRP contains high concentrations of platelets with a large variety of bioactive proteins suggests that PRP can accelerate the tissue regeneration and healing processes.8) Growth factors such as vascular endothelial growth factor, epidermal growth factor (EGF), platelet-derived growth factor (PDGF), insulin-like growth factor, and transforming growth factor-beta (TGF-β) are involved in tissue healing, which are included in α-granules of platelets. It has been revealed that PDGF and TGF-β levels increase as platelet levels become higher, which has been the basis for the clinical use of PRP.9101112) In such processes, growth factors promote 2- to 3-fold tissue healing through angiogenesis, fibroblast activation, elevation of collagen, and glycosaminoglycan syntheses, as well as cell migration and proliferation.131415)
However, little is known about the crucial factors that determine the biological characteristics of PRP. There have been a few studies on the composition of active proteins and expression patterns in PRP itself.161718) Thus, the purpose of this study is to analyze the unknown factors that contribute to tissue healing on the basis of proteomic analysis of proteins in PRP. Since PRP preparation methods and its characteristics are yet to be standardized, its classification and standard also have not been clearly established. Currently, PRP is classified by the kit and the centrifuge used for preparation; level of concentrated components such as platelets and leukocytes; and features of supportive structures for generated products.192021) PRP types practically used can be largely divided into pure or leukocyte poor (LP)-PRP and leukocyte rich (LR)-PRP.22) Thus, to investigate the types and distributions of expressed proteins, we used ACP (Arthrex, Naples, FL, USA) and GPS III (Biomet Biologics Co., Warsaw, IN, USA), which are the most commonly used commercial products for LP-PRP and LR-PRP preparation, respectively, in clinical practice.23) In this study, proteins were identified through two-dimensional gel electrophoresis, followed by gene ontology (GO) analysis.
We collected blood samples from three men who had no underlying disease among the patients who came to our hospital for health checkup. PRP was prepared simultaneously. A total of nine plasma samples (three per person) were obtained (1 session/day) from three men by using the Autologous Conditioned Plasma (ACP) Double Syringe System (Arthrex) for LP-PRP and Gravitational Platelet Separation System (GPS III, Biomet Biologics Co.) for LR-PRP. We conducted this study in compliance with the principles of the Declaration of Helsinki. This study was approved by the Institutional Review Board of Kangnam Sacred Heart Hospital (IRB No. 2012-12-115). All three participants provided informed consent.
Nine milliliters of venous blood was extracted from patients by using a double syringe from the ACP kit and mixed with 1.5 mL of anticoagulant (acid citrate dextrose solution A), which was followed by centrifugation at 380 g for 5 minutes.24) The outer plunger of the double syringe was slowly pressed while the inner plunger was slowly pulled to separate PRP appropriately. Three to five milliliters of generated ACP was moved to the inner syringe to obtain PRP (Fig. 1). During this procedure, care was taken to prevent plasma and erythrocytes from being mixed.
Six milliliters of anticoagulant (acid citrate dextrose solution A) was mixed with 54 mL of venous blood extracted from each patient in each 60-mL syringe in the GPS kit, which was followed by centrifugation at 750 g for 15 minutes.19) Transparent, yellow, platelet-poor plasma at the top of the container was removed by using a syringe in the kit; the remainder was well mixed by centrifugation for 30 seconds, which resulted in about 6 mL of PRP (Fig. 2).
Immediately after collection, the obtained PRP was subjected to a complete blood cell count (CBC) test to investigate the cell composition including leukocytes and platelets. A 1-mL sample of each PRP and blood specimen were analyzed by a hematology analyzer (Siemens ADVIA 2120i; Siemens Healthcare Diagnostics, Erlangen, Germany). The platelet concentration, number of red blood cells, and white blood cell differentiation were calculated. Linearity was (10–1,000) × 103/µL for platelet count; (0.3–7.0) × 106/µL for red blood-cell count; and (0.1–100) × 103/µL for white blood cell count.
Isolated platelets were lysed and the extracted proteins were size-fractionated by one-dimensional-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fifteen micrograms of each protein sample was separated by electrophoresis on a 12% SDS-PAGE gel at 100 V for 1.5 hours (Mini-Protean Bio-Rad, Hercules, CA, USA), stained with Coomassie Blue R-250 (Bio-Rad), and then destained with a destaining solution containing 10% acetic acid and 30% methanol, which was followed by gel imaging. After electrophoresis, the gel was divided into 10 regions by molecular weight, and then fragmented into an 1-mm2 area. Each fragment was subjected to trypsin digestion at 37℃ for 16 hours after cysteine alkylation and reduction of each protein. The digested peptides were eluted by an elution buffer (50 mM ammonium bicarbonate, 50% acetonitrile, and 0.5% trifluoroacetic acid). The collected peptide solution was completely dried by a vacuum dryer (Heto Maxi Dry lyo; Thermo Scientific, Waltham, MA, USA) for 24 hours, which was followed by resuspension in 10 µL of distilled water containing 0.1% formic acid.
LC-MS/MS analysis was performed by an online Nano-LC system and Hybrid Quadrupole-Orbitrap Mass Spectrometry in the Korea Basic Science Institute. Five microliters of peptide samples were resolved by an Ultimate 300 UPLC system (Dionex, Thermo Scientific), which was followed by analysis using the Q-Exactive plus mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray ion source (Dionex, Thermo Scientific). For peptide separation, an Acclaim PepMap RSLC C18 reversed phase (Thermo Scientific) 15 cm × 75 µm column was used in a gradient of 0%–65% ACN at 300 nL/min inflow rate for 2 hours. All MS and MS/MS spectra were acquired in a data-dependent top 10 mode using a Q Exactive plus orbitrap mass spectrometer (Thermo Scientific). The entire MS scan range was 150–2,000 m/z, along with a resolution of 70,000 (m/z 200), an AGC target value of 1 × 106, and a dynamic exclusion of 30 seconds. Fragmentation was performed under higher-energy collisional dissociation (HCD) conditions of either 60 ms in fixed injection times with fast scanning mode in a resolution of 17,500 or 120 ms in fixed injection times with a sensitive method in a resolution of 35,000. The ionization spray voltage was 1.7 kV and capillary temperature was 280℃ along with 25% of HCD collision energy. MS/MS spectra of peptides were subjected to search using MASCOT (Matrix Science, ver. 2.41, https://www.matrixscience.com), in which the protein sequence database of UniprotHuman was used for identification. Mass tolerance and fragmented ions were 10 ppm. Cysteine carbamidomethylation and methionine oxidation were considered as variables of trypsin-digested peptides in MS/MS analysis.
GO analysis was performed to investigate the functions of all identified proteins and classify them according to the AmioGo2 GO database (https://www.AmiGO.geneontology.org).25) To classify the proteins in PRP by function, we determined keywords, focusing on the healing process of damaged tissues such as activation of angiogenesis and fibroblasts as well as increased biosynthesis of collagen and glycosaminoglycan.26)
Thus, wound healing, which comprehensively includes tissue healing in the GO of AmiGo2 was selected as the main keyword for protein classification.25) In AmiGo2, wound healing was defined as a series of events that restores integrity to a damaged tissue after an injury, which included 778 proteins. In addition, angiogenesis involved in wound healing, fibroblast migration, collagen biosynthetic process, glycosaminoglycan biosynthetic process, and glycosaminoglycan binding were selected as keywords that should specifically correspond to the tissue healing process, which was limited only to Homo sapiens. AmiGo2 defined angiogenesis involved in wound healing as blood vessel formation when new vessels emerge from the proliferation of preexisting blood vessels and contribute to the series of events that restore integrity to a damaged tissue after an injury, which included 23 proteins. Fibroblast migration was defined as cell migration that is accomplished by extension and retraction of a fibroblast pseudopodium. A fibroblast is a connective tissue cell, which secretes an extracellular matrix rich in collagen and other macromolecules; it includes 30 proteins. Collagen biosynthetic process was defined as the chemical reactions and pathways resulting in the formation of collagen, any of a group of fibrous proteins of very high tensile strength that form the main component of connective tissue in animals. Collagen is highly enriched in glycine and proline, occurring predominantly as 3-hydroxyproline, which included 47 proteins. Glycosaminoglycan biosynthetic process was defined as the chemical reactions and pathways resulting in the formation of glycosaminoglycan, any of a group of polysaccharides that contain amino sugars, which included 166 proteins. Finally, glycosaminoglycan binding was defined as interacting selectively and noncovalently with any glycan (polysaccharide) containing a substantial proportion of aminomonosaccharide residues, which included 246 proteins.2728)
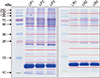
A CBC test was performed immediately after collection to analyze the cell composition of LP-PRP and LR-PRP extracted from whole blood of the three patients. The average cell counts resulting from the test for blood composition were 5.95 (×103/µL) leukocytes and 192 (×103/µL) platelets in whole blood; 1.75 (×103/µL) leukocytes and 371 (×103/µL) platelets were isolated from LP-PRP; and 35 (×103/µL) leukocytes and 719 (×103/µL) platelets were isolated from LR-PRP (Table 1). In other words, platelets were concentrated more than 2- to 3-fold in LP-PRP and 3- to 4-fold in LR-PRP than in whole blood on average. LP-PRP has advantages of being an inexpensive system, which is easy to use, whereas LR-PRP is advantageous to transfer concentrated platelets to patients. For the proteome analysis, a total of nine samples (three samples per each patient) were first separated by electrophoresis, which were divided into 10 fractions. Each sample was subjected to secondary LC-MS/MS analysis in three repetitions (Fig. 3).
As a result, a total of 1,555 proteins were identified, which included proteins identified at least once either in LP-PRP or in LR-PRP. To increase the consistency of samples, only proteins that were identified at least in two of three samples from each patient and were present in at least two patients were classified, which corresponded to 664 of the 1,555 proteins. In detail, the numbers of proteins that were detected at least twice in the three repetitions were 414, 369, and 407 in LP-PRP and 633, 587, and 750 in LR-PRP. Of them, 379 proteins in LP-PRP and 618 proteins in LR-PRP were identified in at least two samples, and 333 proteins were commonly identified in both PRP samples.
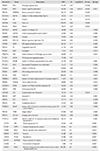
In AmiGo2, 778 proteins corresponded to the keyword, wound healing, and 125 of the 664 identified proteins in the present study were included in this category. Forty-six proteins were analyzed only from LP-PRP, of which eight proteins (FGG, SERPING1, RAP1B, VCL, FLNA, Fbln1, and CLU) were related with wound healing. A total of 285 proteins were analyzed only from LR-PRP, of which 36 proteins (ANXA2, CAPZA1, anxa6, SERPING1, TF, Naca, krt5, MYL12A, WDR1, F9, KNG1, FGG, HSPA5, Gp1bb, Rac2, VCL, C4BPB, FLNA, Arpc4, TUBA4A, Pdia6, CPB2, Aqp1, CFL1, CFD, GNAI2, ITGB3, COL1A2, LAMP2, Gp9, CSRP1, MYL9, RAP1B, CALM1, DMTN, PRTN3) were related to wound healing (Table 2).
Three proteins were included in angiogenesis involved in wound healing, of which one protein (GPX1) and three proteins (GPX1, H3BM21, and ITGB3) were identified in LP-PRP and LR-PRP, respectively (Table 3). A total of two proteins corresponded to fibroblast migration, of which one protein (THBS1) and two proteins (THBS1 and DMTN) were identified in LP-PRP and LR-PRP, respectively (Table 4).
A total of four proteins corresponded to collagen biosynthetic process, of which three proteins (F2, PRDX5, and SERPINF2) and four proteins (F2, PRDX5, SERPINF2, and LTBP1) were identified in LP-PRP and LR-PRP, respectively (Table 5). A total of two proteins corresponded to glycosaminoglycan biosynthetic process, of which one protein (LUM) and two proteins (LUM and CLTC) were identified in LP-PRP and LR-PRP, respectively (Table 6).
A total of 13 proteins corresponded to glycosaminoglycan binding, of which 12 proteins (APOE, IGHM, APOB, CFH, HRG, PF4, SERPIND1, VTN, THBS1, ELANE, LPA, and HABP2) and 13 proteins (APOE, IGHM, APOB, CFH, HRG, PF4, SERPIND1, VTN, THBS1, ELANE, LPA, HABP2, and AZU1) were identified in LP-PRP and LR-PRP, respectively (Table 7).
Since the discovery of PRP, many researchers have reported interesting results on PRP mechanism and its potential in various areas.2) Such research has helped produce results on various proteins and expand the PRP research area. Many preclinical studies and clinical trials also demonstrated promising effects on muscular skeletal diseases.345) However, most of them lacked basic medical evidence, and each study had different procedures for PRP from preparation to injection, making is difficult to compare results of those reports. Thus, this area should be continuously studied along with standardization of biological analysis dependent on the preparation method, activation method, and addition of leukocytes, as well as comparison of various study results.
Despite recent studies on major growth factors in PRP that are related to tissue healing including EGF and PDGF, basic medical studies on their effects still remain to be conducted.811) In addition, the compositions and expression patterns of active proteins in PRP need to be studied. Rapidly identifying and analyzing proteins, which used to be impossible, have become possible, and it became easy to access GO databases. Thus, mutual growths of various science fields have enabled such studies.
Shotgun proteomics can identify a large number of proteins in a short time with a mass spectrometer.2930) This method makes protein samples fragmented into peptides and separates them by using C18 Reversed Phase Chromatography that is connected to the liquid chromatography instrument, along which the sample is simultaneously analyzed. This method requires the establishment of gene databases from complete genome analyses as well as the development of identification programs enabled to perform real-time large-scale analysis using the mass spectrometer.
The present study separated LP-PRP and LR-PRP of human blood through ACP and GPS III, respectively, from which proteins were extracted and resolved by SDS-PAGE electrophoresis for identification. The SDS-PAGE gel was subdivided into 10 fractions, followed by LC-MS/MS analysis, which resulted in the identification of many proteins within a short period of time. To identify the functions of the identified proteins, GO analysis was performed, in which component proteins in PRP were classified by their functions. This resulted in the identification of 125 proteins related to wound healing when both LP-PRP and LR-PRP proteins are combined, which included three proteins for angiogenesis involved in wound healing, two proteins for fibroblast migration, four proteins for collagen biosynthesis process, two proteins for glycosaminoglycan biosynthesis process, and 13 proteins for glycosaminoglycan binding. So new proteins, as well as growth factors that are well-known to be related to tissue healing, were identified.
However, the present study also has limitations such as the small number of subjects (three patients), differences in composition and level of identified proteins between the subjects, and differences in identified proteins even within the same subject. Moreover, a follow-up study should be performed on the interactions between the identified proteins and on degrees of their efficacy in clinical practice.
The results of the present study that is based on the analysis of vast proteomic data of PRP can be used as fundamental data to scientifically identify those factors (both previously known and currently unknown) that are helpful for tissue healing. That is also our goal for a follow-up study. In addition, the examination of the effects of PRP through such protein classification would be the basis for treatment development in new areas based on fundamental medical evidences. Previous proteomic studies were partially focused on platelets such as platelets without stimulation, activated platelets, secretory materials of platelets, platelet membranes, and platelet-derived microparticles. In contrast, the present study performed proteome analysis on PRP itself without activation by addition of either CaCl2 or thrombin, which is clinically significant. In addition, the present study is also meaningful because all proteins within the PRP such as α-granules were investigated without being limited to platelets and classified by their functions in detail, followed by the identification of potential functions of PRP, laying a foundation for basic medical research on tissue healing to be further advanced. PRP has a large potential, which, along with other biological and biochemical advancements, is expected to offer opportunities for more treatments in various areas. In the future, proteomic study would be continued and systematic databases should be constructed, based on which the present study would serve as a foundation for the establishment of basic medical evidence for PRP applications.
Figures and Tables
 | Fig. 1ACP kit of Arthrex. RBC: red blood cell, ACD-A: acid citrate dextrose solution A, PRP: platelet-rich plasma. |
 | Fig. 2GPS kit of Biomet. PPP: platelet-poor plasma, PRP: platelet-rich plasma, RBC: red blood cell, ACD-A: acid citrate dextrose solution A. |
References
1. Ferrari M, Zia S, Valbonesi M, et al. A new technique for hemodilution, preparation of autologous platelet-rich plasma and intraoperative blood salvage in cardiac surgery. Int J Artif Organs. 1987; 10(1):47–50.


2. Dohan Ehrenfest DM, Andia I, Zumstein MA, Zhang CQ, Pinto NR, Bielecki T. Classification of platelet concentrates (Platelet-Rich Plasma-PRP, Platelet-Rich Fibrin-PRF) for topical and infiltrative use in orthopedic and sports medicine: current consensus, clinical implications and perspectives. Muscles Ligaments Tendons J. 2014; 4(1):3–9.



3. de Vos RJ, Weir A, van Schie HT, et al. Platelet-rich plasma injection for chronic Achilles tendinopathy: a randomized controlled trial. JAMA. 2010; 303(2):144–149.


4. Foster TE, Puskas BL, Mandelbaum BR, Gerhardt MB, Rodeo SA. Platelet-rich plasma: from basic science to clinical applications. Am J Sports Med. 2009; 37(11):2259–2272.

5. Mehta S, Watson JT. Platelet rich concentrate: basic science and current clinical applications. J Orthop Trauma. 2008; 22(6):432–438.


7. Orchard J, Seward H. Epidemiology of injuries in the Australian Football League, seasons 1997-2000. Br J Sports Med. 2002; 36(1):39–44.



8. Frechette JP, Martineau I, Gagnon G. Platelet-rich plasmas: growth factor content and roles in wound healing. J Dent Res. 2005; 84(5):434–439.


9. Marx RE, Armentano L, Olavarria A, Samaniego J. rhBMP-2/ACS grafts versus autogenous cancellous marrow grafts in large vertical defects of the maxilla: an unsponsored randomized open-label clinical trial. Int J Oral Maxillofac Implants. 2013; 28(5):e243–e251.

10. Sanchez M, Anitua E, Orive G, Mujika I, Andia I. Platelet-rich therapies in the treatment of orthopaedic sport injuries. Sports Med. 2009; 39(5):345–354.


11. Anitua E, Sanchez M, Orive G, Andia I. The potential impact of the preparation rich in growth factors (PRGF) in different medical fields. Biomaterials. 2007; 28(31):4551–4560.

12. Schultz G, Rotatori DS, Clark W. EGF and TGF-alpha in wound healing and repair. J Cell Biochem. 1991; 45(4):346–352.

13. Anitua E, Andia I, Ardanza B, Nurden P, Nurden AT. Autologous platelets as a source of proteins for healing and tissue regeneration. Thromb Haemost. 2004; 91(1):4–15.


14. Nissen NN, Polverini PJ, Koch AE, Volin MV, Gamelli RL, DiPietro LA. Vascular endothelial growth factor mediates angiogenic activity during the proliferative phase of wound healing. Am J Pathol. 1998; 152(6):1445–1452.


15. Pierce GF, Mustoe TA, Altrock BW, Deuel TF, Thomason A. Role of platelet-derived growth factor in wound healing. J Cell Biochem. 1991; 45(4):319–326.

16. Maynard DM, Heijnen HF, Horne MK, White JG, Gahl WA. Proteomic analysis of platelet alpha-granules using mass spectrometry. J Thromb Haemost. 2007; 5(9):1945–1955.

17. Maguire PB, Wynne KJ, Harney DF, O'Donoghue NM, Stephens G, Fitzgerald DJ. Identification of the phosphotyrosine proteome from thrombin activated platelets. Proteomics. 2002; 2(6):642–648.

18. Garcia A, Prabhakar S, Hughan S, et al. Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins. Blood. 2004; 103(6):2088–2095.


19. Dhurat R, Sukesh M. Principles and methods of preparation of platelet-rich plasma: a review and author's perspective. J Cutan Aesthet Surg. 2014; 7(4):189–197.



20. DeLong JM, Russell RP, Mazzocca AD. Platelet-rich plasma: the PAW classification system. Arthroscopy. 2012; 28(7):998–1009.


21. McCarrel TM, Minas T, Fortier LA. Optimization of leukocyte concentration in platelet-rich plasma for the treatment of tendinopathy. J Bone Joint Surg Am. 2012; 94(19):e143.

22. Harmon KG, Rao AL. The use of platelet-rich plasma in the nonsurgical management of sports injuries: hype or hope? Hematology Am Soc Hematol Educ Program. 2013; 2013:620–626.


23. Fitzpatrick J, Bulsara MK, McCrory PR, Richardson MD, Zheng MH. Analysis of platelet-rich plasma extraction: variations in platelet and blood components between 4 common commercial kits. Orthop J Sports Med. 2017; 5(1):2325967116675272.
24. Noh KC, Liu XN, Zhuan Z, et al. Leukocyte-poor platelet-rich plasma-derived growth factors enhance human fibroblast proliferation in vitro. Clin Orthop Surg. 2018; 10(2):240–247.



25. Carbon S, Ireland A, Mungall CJ, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009; 25(2):288–289.


26. Graziani F, Ivanovski S, Cei S, Ducci F, Tonetti M, Gabriele M. The in vitro effect of different PRP concentrations on osteoblasts and fibroblasts. Clin Oral Implants Res. 2006; 17(2):212–219.


27. O'Neill EE, Brock CJ, von Kriegsheim AF, et al. Towards complete analysis of the platelet proteome. Proteomics. 2002; 2(3):288–305.





 PDF
PDF ePub
ePub Citation
Citation Print
Print








 XML Download
XML Download