1. Schuit FC, Huypens P, Heimberg H, Pipeleers DG. Glucose sensing in pancreatic beta-cells: a model for the study of other glucose-regulated cells in gut, pancreas, and hypothalamus. Diabetes. 2001; 50:1–11.
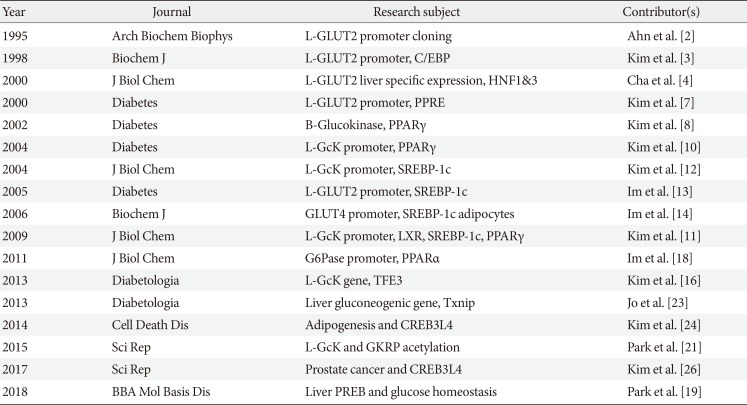
2. Ahn YH, Kim JW, Han GS, Lee BG, Kim YS. Cloning and characterization of rat pancreatic beta-cell/liver type glucose transporter gene: a unique exon/intron organization. Arch Biochem Biophys. 1995; 323:387–396. PMID:
7487103.
3. Kim JW, Ahn YH. CCAAT/enhancer binding protein regulates the promoter activity of the rat GLUT2 glucose transporter gene in liver cells. Biochem J. 1998; 336(Pt 1):83–90. PMID:
9806888.

4. Cha JY, Kim H, Kim KS, Hur MW, Ahn Y. Identification of transacting factors responsible for the tissue-specific expression of human glucose transporter type 2 isoform gene. Cooperative role of hepatocyte nuclear factors 1alpha and 3beta. J Biol Chem. 2000; 275:18358–18365. PMID:
10748140.
5. Cha JY, Kim HS, Kim HI, Im SS, Kim SY, Kim JW, Yeh BI, Ahn YH. Analysis of polymorphism of the GLUT2 promoter in NIDDM patients and its functional consequence to the promoter activity. Ann Clin Lab Sci. 2002; 32:114–122. PMID:
12017192.
6. Nolan JJ, Ludvik B, Beerdsen P, Joyce M, Olefsky J. Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N Engl J Med. 1994; 331:1188–1193. PMID:
7935656.

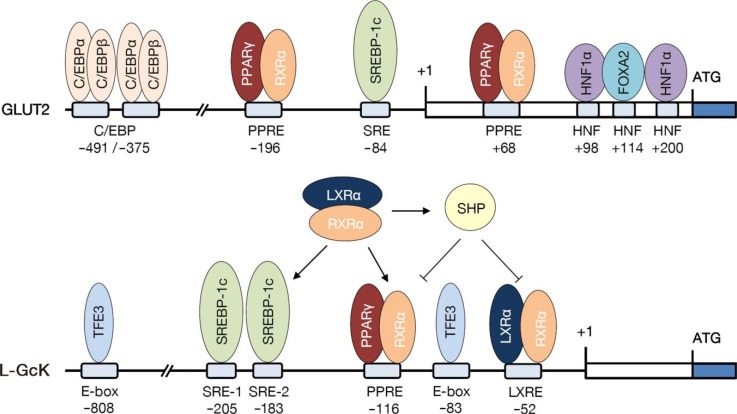
7. Kim HI, Kim JW, Kim SH, Cha JY, Kim KS, Ahn YH. Identification and functional characterization of the peroxisomal proliferator response element in rat GLUT2 promoter. Diabetes. 2000; 49:1517–1524. PMID:
10969836.

8. Kim HI, Cha JY, Kim SY, Kim JW, Roh KJ, Seong JK, Lee NT, Choi KY, Kim KS, Ahn YH. Peroxisomal proliferator-activated receptor-gamma upregulates glucokinase gene expression in beta-cells. Diabetes. 2002; 51:676–685. PMID:
11872666.
9. Kim HI, Ahn YH. Role of peroxisome proliferator-activated receptor-gamma in the glucose-sensing apparatus of liver and beta-cells. Diabetes. 2004; 53(Suppl 1):S60–S65. PMID:
14749267.
10. Kim SY, Kim HI, Park SK, Im SS, Li T, Cheon HG, Ahn YH. Liver glucokinase can be activated by peroxisome proliferator-activated receptor-gamma. Diabetes. 2004; 53(Suppl 1):S66–S70. PMID:
14749268.
11. Kim TH, Kim H, Park JM, Im SS, Bae JS, Kim MY, Yoon HG, Cha JY, Kim KS, Ahn YH. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J Biol Chem. 2009; 284:15071–15083. PMID:
19366697.
12. Kim SY, Kim HI, Kim TH, Im SS, Park SK, Lee IK, Kim KS, Ahn YH. SREBP-1c mediates the insulin-dependent hepatic glucokinase expression. J Biol Chem. 2004; 279:30823–30829. PMID:
15123649.

13. Im SS, Kang SY, Kim SY, Kim HI, Kim JW, Kim KS, Ahn YH. Glucose-stimulated upregulation of GLUT2 gene is mediated by sterol response element-binding protein-1c in the hepatocytes. Diabetes. 2005; 54:1684–1691. PMID:
15919789.

14. Im SS, Kwon SK, Kang SY, Kim TH, Kim HI, Hur MW, Kim KS, Ahn YH. Regulation of GLUT4 gene expression by SREBP-1c in adipocytes. Biochem J. 2006; 399:131–139. PMID:
16787385.

15. Nakagawa Y, Shimano H, Yoshikawa T, Ide T, Tamura M, Furusawa M, Yamamoto T, Inoue N, Matsuzaka T, Takahashi A, Hasty AH, Suzuki H, Sone H, Toyoshima H, Yahagi N, Yamada N. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med. 2006; 12:107–113. PMID:
16327801.

16. Kim MY, Jo SH, Park JM, Kim TH, Im SS, Ahn YH. Adenovirus-mediated overexpression of Tcfe3 ameliorates hyperglycaemia in a mouse model of diabetes by upregulating glucokinase in the liver. Diabetologia. 2013; 56:635–643. PMID:
23269357.

17. Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999; 103:1489–1498. PMID:
10359558.
18. Im SS, Kim MY, Kwon SK, Kim TH, Bae JS, Kim H, Kim KS, Oh GT, Ahn YH. Peroxisome proliferator-activated receptor {alpha} is responsible for the up-regulation of hepatic glucose-6-phosphatase gene expression in fasting and db/db mice. J Biol Chem. 2011; 286:1157–1164. PMID:
21081500.
19. Park JM, Kim MY, Kim TH, Min DK, Yang GE, Ahn YH. Prolactin regulatory element-binding (PREB) protein regulates hepatic glucose homeostasis. Biochim Biophys Acta. 2018; 1864(6 Pt A):2097–2107.

20. Niswender KD, Shiota M, Postic C, Cherrington AD, Magnuson MA. Effects of increased glucokinase gene copy number on glucose homeostasis and hepatic glucose metabolism. J Biol Chem. 1997; 272:22570–22575. PMID:
9278411.

21. Park JM, Kim TH, Jo SH, Kim MY, Ahn YH. Acetylation of glucokinase regulatory protein decreases glucose metabolism by suppressing glucokinase activity. Sci Rep. 2015; 5:17395. PMID:
26620281.

22. Park JM, Kim TH, Bae JS, Kim MY, Kim KS, Ahn YH. Role of resveratrol in FOXO1-mediated gluconeogenic gene expression in the liver. Biochem Biophys Res Commun. 2010; 403:329–334. PMID:
21078299.

23. Jo SH, Kim MY, Park JM, Kim TH, Ahn YH. Txnip contributes to impaired glucose tolerance by upregulating the expression of genes involved in hepatic gluconeogenesis in mice. Diabetologia. 2013; 56:2723–2732. PMID:
24037087.

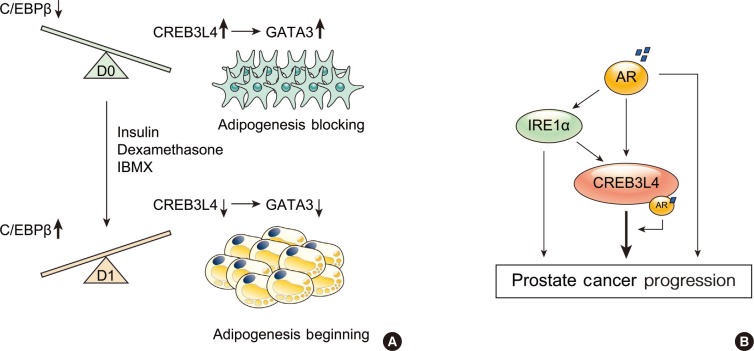
24. Kim TH, Jo SH, Choi H, Park JM, Kim MY, Nojima H, Kim JW, Ahn YH. Identification of Creb3l4 as an essential negative regulator of adipogenesis. Cell Death Dis. 2014; 5:e1527. PMID:
25412305.

25. Kim TH, Park JM, Jo SH, Kim MY, Nojima H, Ahn YH. Effects of low-fat diet and aging on metabolic profiles of Creb3l4 knockout mice. Nutr Diabetes. 2015; 5:e179. PMID:
26302066.

26. Kim TH, Park JM, Kim MY, Ahn YH. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep. 2017; 7:45300. PMID:
28338058.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download