Abstract
The KMT2A (formerly MLL) gene is associated with at least 10% of all cases of acute leukemia. More than 80 translocation partner genes of KMT2A have been discovered to date, six of which have been identified on the long arm of chromosome 17. Among these, the MLLT6 (formerly AF17) gene is located at 17q12 and fuses with the KMT2A gene in rare cases of acute leukemia. We report here a case of AML with a KMT2A/MLLT6 fusion that was confirmed using molecular genetic methods. According to a literature review, this is the first reported case of AML with a KMT2A/MLLT6 fusion in Korea.
The lysine K-specific methyltransferase 2A (KMT2A) gene, formerly known as the MLL gene, is located on chromosome 11q23.3 [1], and its chromosomal rearrangement is implicated in at least 10% of all acute leukemia (AL) cases of various types, including ALL, AML, biphenotypic AL, treatment-related leukemia, and infant leukemia [2]. The KMT2A protein acts as a transcriptional regulator in normal hematopoiesis; AL caused by KMT2A translocation tends to exhibit a more aggressive clinical course than that caused by other etiologies [3].
According to the French-American-British (FAB) classification of AML that is based on cytomorphology and cytochemistry [4], AL caused due to a KMT2A translocation is strongly associated with the monocytic lineage, including an association with M5a in 38.5% of cases, M5b or M4 in 21.2% of cases each, and M2 in 9.6% of cases [5].
More than 80 translocation partner genes (TPGs) of the KMT2A gene have been described to date, many of which have already been cloned and analyzed at the molecular level [2]. Six TPGs located on the long arm of chromosome 17 have been reported: KMT2A/GAS7 (17p13), KMT2A/ACACA (17q21), KMT2A/LASP1 (17q21), KMT2A/MLLT6 (17q21), KMT2A/RARA (17q21), and KMT2A/SEPT9 (17q25) [1].
MLLT6, a PHD finger containing (MLLT6) gene, formerly known as AF17, is located at 17q12, proximal to RARA, which has also been detected in 17q21 [6]. Fusion of the KMT2A and MLLT6 genes is quite rare in AML etiology, as only 2.85% (54 in 1,897) AML cases were confirmed to harbor KMT2A rearrangement [5], and 1.55% (9 in 579) cases of KMT2A-related AML showed a KMT2A/MLLT6 fusion [7]. Herein, we report a case of AML with the KMT2A/MLLT6 fusion resulting in a monoblastic morphology, representing the first such case in Korea, and provide context for the literature on AML showing a KMT2A/MLLT6 fusion reported to date.
A 69-year-old female with a history of hypertension and atherosclerosis after having a fever for one week, accompanied by headache, malaise, and epigastric pain, presented to our hospital. Physical examination showed an afebrile state, without hepatosplenomegaly or abdominal tenderness. A complete blood count revealed a leukocyte count of 6.3×109/L, hemoglobin at 9.6 g/dL, and the platelet count at 5.4×109/L. A peripheral blood smear and manual differential count showed 64% blasts and 26% monocytes. The bone marrow aspirate showed 86.9% large leukemic blasts of an FAB M5a morphology (no faggot cells, Fig. 1A and 1B). Thus, it was diagnosed to be AML. In cytochemistry, the cells were positive for Periodic acid–Schiff stain, and negative for myeloperoxidase, Sudan black B, and dual esterase (<3%). Bone marrow biopsy showed 90% cellularity, and immunohistochemistry showed focal positivity for myeloperoxidase and negativity for terminal deoxynucleotidyl transferase. Immunophenotyping was positive for CD45, CD34, HLA-DR, CD13, CD33, CD64, and CD117, while CD14 was negative.
Chromosomal analysis showed t(11;17)(q23;q21) (Fig. 2), and the reverse transcription (RT)-PCR test (HemaVision-28N, DNA Diagnostics A/S, Risskov, Denmark) was positive for the KMT2A/MLLT6 fusion (Fig. 3). Treatment was initiated with cytarabine and daunorubicin. After one month of therapy, follow-up bone marrow aspiration showed a hypocellular marrow with 2.49% blasts. No chromosomal analysis was conducted at this point due to the lack of mitotic cells; however, FISH analysis (KMT2A Break-apart, Cytocell, Cambridge, UK) identified a rearrangement of the KMT2A gene in 2.5% of interphase cells (Fig. 4). No further chemotherapy was administered due to neutropenia. Four months after the initial diagnosis, the leukemia relapsed, and the patient ultimately died from septic shock. Next-generation sequencing (NGS) of the frozen RNA sample (taken at initial diagnosis) and a gene panel test which can identify the gene fusion (Oncomine Myeloid Research Assay, Thermo Scientific Inc., Waltham, MA, USA) were negative for the fusion gene.
A total of 13 cases of AML with the KMT2A/MLLT6 fusion have been reported to date [2,8,9,10,11,12,13] (Table 1). Prasad et al. [8] reported the first such case involving the fusion of KMT2A exon 5 and an unknown exon of MLLT6 based on RT-PCR and the Sanger sequencing method. Suzukawa et al. [9] found a breakpoint at KMT2A exon 6 and MLLT6 exon 9, and confirmed the KMT2A rearrangement by FISH analysis. Moore et al. [10] used a gene-specific probe to confirm the KMT2A/MLLT6 fusion in FISH analysis, whereas Grossman et al. [12] applied NGS for identification of this fusion for the first time with the pyrosequencer (454 FLX, Roche Diagnostics, Rotkreuz, Switzerland).
The MLLT6 gene is located on chromosome 17q12, but karyotype analysis has provided diverse results such as 17q21 [9–12 and present case] or 17q12-21 [2] in previous literature. This discrepancy is likely due to resolution differences in the karyotype analyses [10] or reflects the molecular heterogeneity of AML patients [11].
In our case, we used NGS sequence analysis (S5 XL, Thermo Scientific Inc.) and a gene panel test for hematologic malignancy (Oncomine Myeloid Research Assay, Thermo Scientific Inc.) to identify the gene fusion, but were unable to find the gene fusion using these methods. KMT2A is included in the RNA fusion gene list of the assay [14], but the sample was stored for about two years at −20℃. Since ribonuclease can remain active at this temperature [15], the low sample quality might explain this negative result.
This is the first case of KMT2A/MLLT6 fusion in a patient with AML reported in Korea. Kim et al. [16] reported a case of AML M5 with t(11;17)(q23;21) in 2006. In this case, FISH analysis revealed that the breakpoint in chromosome 17 was proximal to the RARA gene, but the authors did not perform a RT-PCR test to confirm the KMT2A gene rearrangement. Several cases of AML have been reported to be associated with rearrangement of the KMT2A gene with the region proximal to the RARA gene [17,18]; however, these might not involve the KMT2A/MLLT6 fusion, because two other TPGs (besides MLLT6 and RARA) are located in that region [1] and should thus be confirmed by more sensitive assays such as RT-PCR or NGS.
Unlike the FAB classification based on morphological findings, the WHO classification uses cytogenetic characteristics for classification. AML with the KMT2A/MLLT6 fusion is not yet accepted as a full entity in the WHO classification [19], but the aggressive disease course shown in the present case emphasizes the importance of molecular genetic assays in addition to morphology, cytochemistry and cytogenetic analyses. King et al. [20] reported 12% (7/57) of cases of acute leukemia which had translocation were positive using the RT-PCR method and negative using the conventional cytogenetic assay. Therefore, for patients newly diagnosed as having AL, RT-PCR or FISH should be performed to find chromosomal aberrations not detected by conventional cytogenetic assays.
Figures and Tables
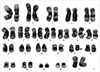 | Fig. 1Peripheral blood and bone marrow smears of the patient. (A) A peripheral blood smear shows monoblasts with abundant cytoplasm and prominent nucleoli (Wright stain, ×400). (B) A bone marrow aspiration smear shows uniform proliferation of monoblasts which have round/folded nuclear and abundant cytoplasm (Wright stain, ×400). |
 | Fig. 2 |
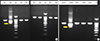 | Fig. 3(A) A reverse transcription-PCR assay shows a band in the second lane (M2). (B) Split-out image shows a band (approximately 600 base pairs) in lane 2B, which indicates KMT2A/MLLT6 fusion transcripts. |
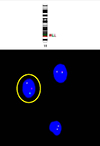 | Fig. 4FISH analysis of a follow-up bone marrow sample with dual color (green and red) probes of KMT2A. The circled cell shows one yellow, one green and one red signal, which indicate KMT2A rearrangement (in 5/200 interphase cells, reference range: < 2%). |
References
1. Huret JL. MLL (myeloid/lymphoid or mixed lineage leukemia). Atlas Genet Cytogenet Oncol Haematol. 2006; 10:83–87.

2. Burillo-Sanz S, Morales-Camacho RM, Caballero-Velázquez T, Carrillo E, Sánchez J, Pérez-López O, et al. MLL-rearranged acute myeloid leukemia: Influence of the genetic partner in allo-HSCT response and prognostic factor of MLL 3′ region mRNA expression. Eur J Haematol. 2018; 100:436–443.

3. Deqing Hu. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016; 30:2021–2041.

4. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985; 103:620–625.

5. Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003; 102:2395–2402.

6. Huret JL. AF17 (ALL1 fused gene from chromosome 17). Atlas Genet Cytogenet Oncol Haematol. 1997; 1:53.

7. Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013; 27:2165–2176.
8. Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H, et al. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc Natl Acad Sci USA. 1994; 91:8107–8111.

9. Suzukawa K, Shimizu S, Nemoto N, Takei N, Taki T, Nagasawa T. Identification of a chromosomal breakpoint and detection of a novel form of an MLL-AF17 fusion transcript in acute monocytic leukemia with t(11;17)(q23;q21). Int J Hematol. 2005; 82:38–41.

10. Moore SD, Strehl S, Dal Cin P. Acute myelocytic leukemia with t(11;17)(q23;q12-q21) involves a fusion of MLL and AF17. Cancer Genet Cytogenet. 2005; 157:87–89.

11. Strehl S, König M, Meyer C, Schneider B, Harbott J, Jäger U, et al. Molecular dissection of t(11;17) in acute myeloid leukemia reveals a variety of gene fusions with heterogeneous fusion transcripts and multiple splice variants. Genes Chromosomes Cancer. 2006; 45:1041–1049.

12. Grossmann V, Kohlmann A, Klein HU, Schindela S, Schnittger S, Dicker F, et al. Targeted next-generation sequencing detects point mutations, insertions, deletions and balanced chromosomal rearrangements as well as identifies novel leukemia-specific fusion genes in a single procedure. Leukemia. 2011; 25:671–680.

13. De Braekeleer E, Meyer C, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, et al. Identification of MLL partner genes in 27 patients with acute leukemia from a single cytogenetic laboratory. Mol Oncol. 2011; 5:555–563.

14. ThermoFisher Scientific. Oncomine myeloid research assay. last visited on Jan 2019. https://www.thermofisher.com/kr/ko/home/clinical/preclinical-companion-diagnostic-development/oncomine-oncology/oncomine-myeloid-research-assay.html.
15. Ma S, Huang Y, van Huystee RB. Improved plant RNA stability in storage. Anal Biochem. 2004; 326:122–124.

16. Kim KE, Kim SH, Han JY. Acute monocytic leukemia with t(11;17)(q23;q21) involving a rearrangement of mixed lineage leukemia gene. Korean J Lab Med. 2006; 26:329–333.

17. Reeves BR, Kempski H, Jani K, Borrow J, Howe K, Solomon E, et al. A case of acute monocytic leukemia with t(11;17) involving a rearrangement of MLL-1 and a region proximal to the RARA gene. Cancer Genet Cytogenet. 1994; 74:50–53.

18. Douet-Guilbert N, Morel F, Le Bris MJ, Herry A, Morice P, Bourquard P, et al. Rearrangement of the MLL gene in acute myeloblastic leukemia: report of two rare translocations. Cancer Genet Cytogenet. 2005; 157:169–174.

19. Swerdlow SH, Campo E, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon, France: IARC;2017.
20. King RL, Naghashpour M, Watt CD, Morrissette JJ, Bagg A. A comparative analysis of molecular genetic and conventional cytogenetic detection of diagnostically important translocations in more than 400 cases of acute leukemia, highlighting the frequency of false-negative conventional cytogenetics. Am J Clin Pathol. 2011; 135:921–928.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download