1. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016; 6:34921.

2. Dennie C, Thornhill R, Sethi-Virmani V, Souza CA, Bayanati H, Gupta A, et al. Role of quantitative computed tomography texture analysis in the differentiation of primary lung cancer and granulomatous nodules. Quant Imaging Med Surg. 2016; 6:6–15.


4. Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, et al. Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. 2019; 290:783–792.


5. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014; 5:4006.

6. McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015; 27:15–26.


7. Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol. 2012; 22:796–802.


8. Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology. 2013; 266:326–336.


9. Win T, Miles KA, Janes SM, Ganeshan B, Shastry M, Endozo R, et al. Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer. Clin Cancer Res. 2013; 19:3591–3599.


10. Fried DV, Tucker SL, Zhou S, Liao Z, Mawlawi O, Ibbott G, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2014; 90:834–842.


11. Cherezov D, Goldgof D, Hall L, Gillies R, Schabath M, Müller H, et al. Revealing tumor habitats from texture heterogeneity analysis for classification of lung cancer malignancy and aggressiveness. Sci Rep. 2019; 9:4500.

12. Cook GJ, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013; 54:19–26.


13. Cook GJ, O'Brien ME, Siddique M, Chicklore S, Loi HY, Sharma B, et al. Non-small cell lung cancer treated with erlotinib: heterogeneity of
18F-FDG uptake at PET—Association with treatment response and prognosis. Radiology. 2015; 276:883–893.

15. Poonyagariyagorn H, Mazzone PJ. Lung cancer: preoperative pulmonary evaluation of the lung resection candidate. Semin Respir Crit Care Med. 2008; 29:271–284.


16. Wu MT, Chang JM, Chiang AA, Lu JY, Hsu HK, Hsu WH, et al. Use of quantitative CT to predict postoperative lung function in patients with lung cancer. Radiology. 1994; 191:257–262.


17. Humphries SM, Yagihashi K, Huckleberry J, Rho BH, Schroeder JD, Strand M, et al. Idiopathic pulmonary fibrosis: data-driven textural analysis of extent of fibrosis at baseline and 15-month follow-up. Radiology. 2017; 285:270–278.


18. Maldonado F, Moua T, Rajagopalan S, Karwoski RA, Raghunath S, Decker PA, et al. Automated quantification of radiological patterns predicts survival in idiopathic pulmonary fibrosis. Eur Respir J. 2014; 43:204–212.


19. Moon JW, Bae JP, Lee HY, Kim N, Chung MP, Park HY, et al. Perfusion- and pattern-based quantitative CT indexes using contrast-enhanced dual-energy computed tomography in diffuse interstitial lung disease: relationships with physiologic impairment and prediction of prognosis. Eur Radiol. 2016; 26:1368–1377.

20. Park HJ, Lee SM, Song JW, Lee SM, Oh SY, Kim N, et al. Texture-based automated quantitative assessment of regional patterns on initial CT in patients with idiopathic pulmonary fibrosis: relationship to decline in forced vital capacity. AJR Am J Roentgenol. 2016; 207:976–983.


21. Yoon RG, Seo JB, Kim N, Lee HJ, Lee SM, Lee YK, et al. Quantitative assessment of change in regional disease patterns on serial HRCT of fibrotic interstitial pneumonia with texture-based automated quantification system. Eur Radiol. 2013; 23:692–701.


22. Yang X, Pan X, Liu H, Gao D, He J, Liang W, et al. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis. 2018; 10:Suppl 7. S807–S819.

23. Zhong Y, Yuan M, Zhang T, Zhang YD, Li H, Yu TF. Radiomics approach to prediction of occult mediastinal lymph node metastasis of lung adenocarcinoma. AJR Am J Roentgenol. 2018; 211:109–113.


24. Bayanati H, Thornhill RE, Souza CA, Sethi-Virmani V, Gupta A, Maziak D, et al. Quantitative CT texture and shape analysis: can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol. 2015; 25:480–487.


25. Andersen MB, Harders SW, Ganeshan B, Thygesen J, Torp Madsen HH, Rasmussen F. CT texture analysis can help differentiate between malignant and benign lymph nodes in the mediastinum in patients suspected for lung cancer. Acta Radiol. 2016; 57:669–676.


26. Coroller TP, Agrawal V, Huynh E, Narayan V, Lee SW, Mak RH, et al. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol. 2017; 12:467–476.


27. Li H, Becker N, Raman S, Chan TC, Bissonnette JP. The value of nodal information in predicting lung cancer relapse using 4DPET/4DCT. Med Phys. 2015; 42:4727–4733.


28. Gevaert O, Echegaray S, Khuong A, Hoang CD, Shrager JB, Jensen KC, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep. 2017; 7:41674.

29. Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol. 2016; 26:32–42.


30. Yoon HJ, Sohn I, Cho JH, Lee HY, Kim JH, Choi YL, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine (Baltimore). 2015; 94:e1753.

31. Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, et al. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology. 2018; 286:307–315.


33. Padhani AR, Miles KA. Multiparametric imaging of tumor response to therapy. Radiology. 2010; 256:348–364.


34. Lee HY, Jeong JY, Lee KS, Yi CA, Kim BT, Kang H, et al. Histopathology of lung adenocarcinoma based on new IASLC/ATS/ERS classification: prognostic stratification with functional and metabolic imaging biomarkers. J Magn Reson Imaging. 2013; 38:905–913.


35. Kim J, Ryu SY, Lee SH, Lee HY, Park H. Clustering approach to identify intratumour heterogeneity combining FDG PET and diffusion-weighted MRI in lung adenocarcinoma. Eur Radiol. 2019; 29:468–475.


36. Even AJG, Reymen B, La Fontaine MD, Das M, Mottaghy FM, Belderbos JSA, et al. Clustering of multi-parametric functional imaging to identify high-risk subvolumes in non-small cell lung cancer. Radiother Oncol. 2017; 125:379–384.


37. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014; 9:e102107.

38. Lassen BC, Jacobs C, Kuhnigk JM, van Ginneken B, van Rikxoort EM. Robust semi-automatic segmentation of pulmonary subsolid nodules in chest computed tomography scans. Phys Med Biol. 2015; 60:1307–1323.


40. Devaraj A, van Ginneken B, Nair A, Baldwin D. Use of volumetry for lung nodule management: theory and practice. Radiology. 2017; 284:630–644.


41. Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017; 35:18–31.


42. Trebeschi S, van Griethuysen JJM, Lambregts DMJ, Lahaye MJ, Parmar C, Bakers FCH, et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep. 2017; 7:5301.

44. Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016; 6:23428.

45. Yip S, McCall K, Aristophanous M, Chen AB, Aerts HJ, Berbeco R. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PLoS One. 2014; 9:e115510.

46. Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol. 2015; 8:524–534.

47. Du Q, Baine M, Bavitz K, McAllister J, Liang X, Yu H, et al. Radiomic feature stability across 4D respiratory phases and its impact on lung tumor prognosis prediction. PLoS One. 2019; 14:e0216480.

48. Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One. 2016; 11:e0164924.

49. Lo P, Young S, Kim HJ, Brown MS, McNitt-Gray MF. Variability in CT lung-nodule quantification: effects of dose reduction and reconstruction methods on density and texture based features. Med Phys. 2016; 43:4854.

50. Solomon J, Mileto A, Nelson RC, Roy Choudhury K, Samei E. Quantitative features of liver lesions, lung nodules, and renal stones at multi-detector row CT examinations: dependency on radiation dose and reconstruction algorithm. Radiology. 2016; 279:185–194.


52. Alahmari SS, Cherezov D, Goldgof D, Hall L, Gillies RJ, Schabath MB. Delta radiomics improves pulmonary nodule malignancy prediction in lung cancer screening. IEEE Access. 2018; 6:77796–77806.

54. van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014; 15:3221–3245.
55. Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): a new radiomics descriptor. Sci Rep. 2016; 6:37241.

56. Incoronato M, Aiello M, Infante T, Cavaliere C, Grimaldi AM, Mirabelli P, et al. Radiogenomic analysis of oncological data: a technical survey. Int J Mol Sci. 2017; 18:E805.

57. Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017; 28:1191–1206.


58. Kim ST, Lee JH, Lee H, Ro YM. Visually interpretable deep network for diagnosis of breast masses on mammograms. Phys Med Biol. 2018; 63:235025.

59. Tang Z, Chuang KV, DeCarli C, Jin LW, Beckett L, Keiser MJ, et al. Interpretable classification of Alzheimer's disease pathologies with a convolutional neural network pipeline. Nat Commun. 2019; 10:2173.

60. Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016; 35:1299–1312.


62. Ru Zhao Y, Xie X, de Koning HJ, Mali WP, Vliegenthart R, Oudkerk M. NELSON lung cancer screening study. Cancer Imaging. 2011; 11 Spec No A:S79–S84.

63. Sanchez-Salcedo P, Berto J, de-Torres JP, Campo A, Alcaide AB, Bastarrika G, et al. Lung cancer screening: fourteen year experience of the Pamplona early detection program (P-IELCAP). Arch Bronconeumol. 2015; 51:169–176.



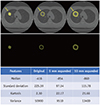
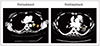
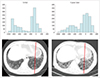


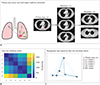





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download