INTRODUCTION
Cardiovascular disease (CVD) is the top cause of death worldwide. Several risk factors for CVD have been identified, including cholesterol levels, hypertension, and others. However, even when combined, these known risk factors do not completely predict incident CVD. Clearly, the identification of novel biomarkers associated with CVD would yield an improved understanding of disease etiology, aid in the prediction of incident disease, and point toward new therapeutic targets.
In addition to cholesterol and triglycerides, ceramides are another type of bioactive lipid species that are of interest as potential markers of CVD risk. The identification of novel biomarkers associated with these diseases would improve our understanding of the etiology of CVD, assist in the prediction of incident CVD, and shed light on novel potential therapeutic targets. Broad inhibition of ceramide synthesis prevents diet- and glucocorticoid-induced insulin resistance in mice, thus implicating ceramides in the promotion of metabolic and CVD.
12 Moreover, in several murine models of metabolic cardiomyopathy, and in humans, total cardiac ceramide accumulation is associated with cardiac dysfunction.
345 Recently, however, studies by our group
6 have demonstrated that plasma levels of specific very-long-chain molecular species of ceramides (C24:0 and C22:0) are associated with a decreased risk of coronary heart disease (CHD), heart failure (HF), and/or all-cause mortality. For example, the relative risks of CHD, HF, and all-cause mortality for each 3-unit increase in plasma C24:0 ceramide levels were 0.79, 0.75, and 0.78, respectively, even after adjusting for standard CVD risk factors including blood cholesterol. Other groups have demonstrated that a lower risk of CVD events was associated with increased plasma C24:0 ceramide and/or C22:0 ceramide concentrations in patients with known CHD.
78
There is both a
de novo and a salvage pathway for sphingolipid and ceramide production, and specific ceramide synthases create ceramides of specific chain lengths. Both ceramide synthase 2 and 3 can generate the very-long-chain C22:0 and C24:0 species.
9
However, the genomic links between these species are incompletely understood. We hypothesized that specific loci would be associated with these 2 important complex lipid species. To test this hypothesis, we leveraged existing genome-wide scans to examine variants associated with C22:0 and C24:0 ceramide concentrations. Our primary aim was to identify variants that met genome-wide significance. However, with an exploratory, hypothesis-generating aim, we lowered our threshold for significance (to p<1E-5) and used pathway analyses to identify which genes had the strongest potential causal association and would be suitable for further analyses.
DISCUSSION
Ceramides are complex lipids composed of sphingosine and a fatty acid of varying acyl chain length. Ceramides have pleiotropic effects. They are found in cell membranes and can affect cell signaling, apoptosis, differentiation, senescence, and proliferation, as well as hormonal and stress responses. Studies of ceramides have shown that high total ceramide concentrations in plasma and/or tissue are related to increased insulin resistance, CVD, and cardiovascular death in patients with HF.
5131415 However, higher concentrations of some very-long-chain ceramides in the plasma predict a decreased risk of CHD, HF, and/or all-cause mortality.
68 We hypothesized that plasma C22:0 and C24:0 ceramide concentrations would be heritable and would be associated with genetic variants.
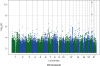
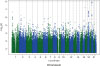
In this study, we demonstrated the heritability of plasma concentrations of C22:0 and C24:0 ceramides. We also showed that 19 SNPs on chromosome 20 were associated with plasma C22:0 ceramide concentrations and that 9 SNPs on chromosome 20 were associated with plasma C24:0 ceramide concentrations at a GWAS level of significance (p≤5E-8). The SNPs were in strong linkage disequilibrium with each other, and when a conditional analysis on the lead SNP was performed (conditioned on rs4814175 for C22:0 ceramide and rs168622 for C24:0 ceramide), the effects of the other genome-wide significant variants were attenuated to non-significance, indicating that the SNPs captured the same genetic effect. All of the SNPs were within 60 kb of the gene encoding SPTLC3.
SPTLC3 is a subunit of the enzyme serine palmitoyl transferase (SPT), which catalyzes the rate-limiting step of
de novo synthesis of the sphingoid backbone of sphingolipids, which are precursors for ceramide synthesis. SPT condenses a fatty acyl-coenzyme A and serine to create a sphingoid base.
16 SPT is composed of 1 or more heterodimers, including the SPTLC1 subunit, which links the SPT enzyme to the membrane, and a catalytic subunit (e.g., SPTLC3 or SPTLC2).
16 SPTLC3 is highly expressed in the myocardium.
16 The SPTLC3 subunit enables the SPT enzyme to generate either d16 or d18 sphingoid bases, which are precursors in ceramide production.
16 The specific sphingoid base of the ceramides measured in this study was 18:1. Different sphingoid base lengths can cause different effects. For example, d16 bases enhance cell death pathways.
16 In addition, different acyl chain lengths appear to have differential effects on disease pathways. Prior investigators have linked certain ceramide species (C18:1, C18:0, C20:0, and C24:1 dihydroceramide), with conditions related to CVD, including diabetes, insulin resistance, and increased plasma levels of inflammatory mediators, such as tumor necrosis factor alpha.
1718 One small study has also linked C22:0 levels with insulin resistance.
17
Genetic polymorphisms in
SPTLC3 associated with metabolic phenotypes have also been previously reported. A recent GWAS investigating 163 metabolic traits revealed that 9 SNPs, including the
SPTLC rs168622 variant, were associated with the variability of serum levels of different metabolites and serum metabolite ratios (thought to be representative of enzymatic reaction rates).
19 Specifically, it was found that the
SPTLC rs168622 G variant was associated with higher C24:1 concentrations and lower C24:0/C24:1 concentrations.
19 Furthermore, as mentioned above, higher C24:1 levels have been associated with insulin resistance. It is challenging to explain exactly how changing 1 subunit (SPTLC3) in a multi-complex enzyme (SPT) would result in these and other changes in the concentrations of ceramides of different fatty acyl chain lengths. Changes in SPTLC3 may alter SPT activity or cause compensatory changes in the expression of other subunits or activity that may lead to different chain length preferences. In addition, multiple biochemical pathways (e.g., the salvage pathway in addition to the
de novo pathway) lead to ceramide synthesis. Precisely how changing the efficiency of a single pathway will alter the balance of acyl chains is complex and not readily predictable, as different pathways may lead to preferences for different acyl chain incorporation, and there could be changes in the expression or activity of other inputs to ceramide synthesis. Other investigators have shown that carriers of the
SPTLC3 rs168622 GG variant had lower relative mRNA expression of
SPTLC3 and lower total hepatic lipid content.
20 Since the
SPTLC3 rs168622 GG variant leads to lower mRNA expression of
SPTLC3, it is likely that those individuals have less SPTLC3-mediated generation of d16 (and d18) bases, leading to lower levels of ceramides composed of d16 bases, which are linked with cell death. Taken together, these prior observations regarding sphingoid base and/or fatty acyl chain effects of
SPTLC3 SNPs lend biological plausibility to our findings and suggest potential molecular mechanisms connecting our genomic findings with CVD and its risk factors.
As part of our hypothesis-generating aim, we identified 2 SNPs and 1 pathway that did not reach genome-wide significance, but had a false discovery rate of less than 20%: rs79935880 (
p=8.05E-7), ENSG00000138107 (
p=2.42E-6) and REACTOME_SIGNALING_BY_ROBO_RECEPTOR (
p=2.28E-5). The rs79935880 SNP is a variant of the
ANKRD30A gene, which encodes a transcription factor that is only expressed in breast and testicular tissue. ENSG00000138107 is located near
ACTR1A, which encodes actin related protein 1A, a subunit of dynactin. REACTOME_SIGNALING_BY_ROBO_RECEPTOR is a set of 30 genes that are involved in signaling by the Robo receptor; these genes encode transmembrane receptors that regulate axonal guidance and cell migration and are important in development. While the biological plausibility of rs79935880 and REACTOME_SIGNALING_BY_ROBO_RECEPTOR may not be immediately apparent, ENSG00000138107 has obvious potential biological relevance. Ceramides are signaling molecules that are upregulated in response to several physiological stressors. The specific effects of these complex lipids appear to relate to the type of tissue they are in, as well as their acyl chain length.
21 Ceramides have a variety of effects, including pro-apoptosis, pro-mitophagy, pro-survival, and pro-autophagy.
22 Specific ceramide species have been demonstrated to promote programmed cell death through a cascade of events involving ceramide-mediated channels and a loss of mitochondrial outer membrane integrity that facilitate the release of cytochrome c and other pro-apoptotic proteins into the cytoplasm, ultimately resulting in mitophagy and mitochondria-induced apoptosis.
2324 In contrast, C24:0 ceramide interferes with this process and has predominantly anti-apoptotic effects.
25 Given that dynactin is involved in transport between the endoplasmic reticulum and Golgi apparatus, as well as the formation and movement of lysosomes and endosomes, this gene may be an important component of the pathway(s) that are influenced by C22:0 and C24:0 ceramides, a finding that may provide further insight into the mechanisms underlying the effects of ceramides.
Our results validate and extend the findings of previous investigators. Hicks and colleagues assessed whether a GWAS analysis could identify variants associated with the concentrations of 33 circulating sphingolipids (24 sphingomyelins and 9 ceramides) in 5 large, presumed to be healthy, European populations.
26 The 20p12.1 locus was found to have the strongest association with serum ceramide concentrations. Although no SNP reached genome-wide significance in any of the 5 populations, rs680379 reached genome-wide significance for its associations with both C22:0 and C24:0 ceramide concentrations in a meta-analysis of all 5 populations. In contrast, when Lemaitre and colleagues
27 investigated whether a GWAS analysis could identify variants associated with the total plasma phospholipid and erythrocyte levels of 3 saturated, very-long-chain fatty acids (20:0, 22:0, and 24:0, which included ceramides with these fatty acid acyl chains—C20:0, C22:0, and C24:0—as well as other lipid moieties) in a meta- analysis of 7 cohorts comprising 10,129 individuals of European ancestry, no genome-wide significant associations were found with circulating levels of 22:0 or 24:0 fatty acids. However, C20:0 ceramide concentrations were associated with 7 variants in strong linkage disequilibrium with each other on chromosome 20p12.1; the most significant variant was rs680379.
27 Our findings support those of Hicks and colleagues.
26 We also observed that the rs680379 variant reached genome-wide significance for its associations with both C22:0 and C24:0 ceramide concentrations. Our findings in the FHS Offspring Cohort provide important validation and confirmation that this locus is an important genetic determinant of C22:0 and C24:0 ceramide concentrations.
The identification of SNPs that are associated with plasma ceramide concentrations is important for several reasons. Patients' SPTLC3 genotype status should be considered, and potentially adjusted for, when assessing plasma ceramide concentrations and determining individual risk and prognosis. We speculate that in the future, use of genotype-guided, precision medicine approaches might identify specific patient groups that could benefit from unique, targeted therapeutic interventions.
Our study should be interpreted in the context of several potential limitations. First, the FHS Offspring Cohort contains white individuals of European ancestry. Thus, it is unclear whether these results are generalizable to individuals of different racial or ethnic groups. Second, our conditional analyses that identified rs79935880, ENSG00000138107, and REACTOME_SIGNALING_BY_ROBO_RECEPTOR as potentially significant should be considered as exploratory, as they have not been validated in an independent cohort. However, these findings, and particularly the association with ACTR1A, are intriguing as they have biological plausibility and may lead to insights into novel pathways and therapeutics that could benefit specific populations. In addition, associations (between SNPs related to SPTLC3 and very-long-chain ceramides, as well as between very-long-chain ceramides and CVD) do not prove causation. Future studies are needed to further investigate the possible causative roles of genes and specific ceramides for disease. Lastly, the possible effects of the specific SNPs identified as being significantly related to plasma C22:0 and C24:0 ceramides on other ceramide species is beyond the scope of this study.
In summary, we have identified SNPs on chromosome 20 associated with plasma C22:0 and C24:0 ceramide concentrations in individuals from the FHS Offspring Cohort. These SNPs are near SPTLC3, the gene encoding SPTLC3, which catalyzes the rate-limiting step of de novo sphingolipid synthesis. Our results are biologically plausible and suggest that SPTLC3 may be a novel therapeutic target for C24:0 and C22:0 ceramide modulation.









 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download