Abstract
In order to investigate the antioxidant effect of alkylhydroxide peroxidase (ahpC) of Helicobacter pylori (H. pylori) 26695, an ahpC-deficient mutant (H. pylori 26695 ahpC::cat) was generated. ahpC-deficient mutant was grown slowly at lower pressure of oxygen (5% oxygen) compared to the H. pylori 26695. Whole cell proteins isolated form H. pylori 26695 and H. pylori 26695 ahpC::cat were analyzed by MALDI-TOF and tandem-MS. The expression of 15 proteins, including Ppa, HypB, GrpE, Elp, RecA, GroES, Mda66, RibE, NapA, GlnA, BioB, TrxB, Tsf, FumC and Icd, was more than doubled in H. pylori 26695 ahpC::cat. Production of 10 proteins such as UreG, FabE, Adk, Pnp, OorC, AtpA, AtpD, Nqq3, Pfr, and TagD decreased below 50% in H. pylori 26695 ahpC::cat compared to the H. pylori 26695. In microarray analysis, 9 genes including sul1, amiE, frxA, fecA, hyuA, and katA increased in transcription level in H. pylori 26695 ahpC::cat compared to H. pylori 26695. A total of 24 genes, including flaB, protein kinase C inhibitor, cag16, pabC, and sabA, reduced in transcription. 27 genes, including HP0889, showed common expression changes in ahpC, katA, and sodB-deficient mutations. As a result of this study, there were not many genes whose expression was commonly changed by the deletion of each of the three major antioxidant enzymes of H. pylori. These results showed the functions and regulation of the three antioxidant enzymes were different in H. pylori.
Figures and Tables
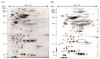 | Figure 22D Separation of whole cell proteins of the wild-type (H. pylori 26695) (A), H. pylori 26695 ahpC::cat (B). The proteins were separated on an IPG strip of pH 5.0-8.0 and subsequently on a 12.5% SDS-PAGE and then detected by silver staining. The original gel size was 18×20×0.15. A) Wild type. B) H. pylori 26695 ahpC::cat. The protein spots corresponding to AhpC are marked with rectangle. kDa shows the molecular mass (indicated on left). |
 | Figure 3Growth curve of H. pylori 26695 and H. pylori 26695 ahpC::cat at thin layer culture condition. |
Table 2
List of up- or down-regulated proteins in ahpC-deficient isogenic mutant compared to H. pylori 26695 by using tandem ms.

Table 3
List of up- or down-regulated proteins in ahpC-deficient isogenic mutant compared to H. pylori 26695 by using MALDI-TOF.

ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2016R1A2B1015791).
References
1. Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 1:1311–1354.

2. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994; 61:1–241.
3. Campylobacter-like organisms in the stomach of patients and healthy individuals. Lancet. 1984; 1:1348–1349.
4. Rhee KH, Cho MJ, Kim JB, Choi SK, Park CK, Kim YC, et al. A prospective study on Campylobacter pylori isolated from patients of gastroduodenal inflammatory conditions. J Korean Soc Microbiol. 1988; 23:9–16.
5. Petersen AM, Krogfelt KA. Helicobacter pylori: an invading microorganism? A review. FEMS Immunol Med Microbiol. 2003; 36:117–126.
6. Akada JK, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol. 2000; 36:1071–1084.

7. Higashi H, Tsutsumi , Muto S. SHP-2 tyrosine phosphatase as an intacellular target of Helicobacter pylori CagA protein. Science. 2002; 295:683–686.

8. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–547.

9. Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999; 397:176–180.

11. Cho MJ, Lee SG, Lee KH, Song JY, Lee WK, Baik SC, et al. Comparison of gene expression patterns between Helicobacter pylor 26695 and its superoxide dismutase isogenic mutant. J Bacteriol Virol. 2013; 43:279–289.

12. Kang HL, Lee SG, Park JS, Song JY, Cho MJ, Baik SC, et al. Proteome Analysis of a Catalase-deficient Isogenic Mutant of Helicobacter pylori 26695. J Bacteriol Virol. 2014; 44:177–187.

13. Seyler RW Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001; 69:4034–4040.

14. Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006; 61:847–860.

15. Joo JS, Park KC, Song JY, Kim DH, Lee KJ, Kwon YC, et al. A thin-layer liquid culture technique for the growth of Helicobacter pylori. Helicobacter. 2010; 15:295–302.

16. O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975; 250:4007–4021.
17. Wang Y, Taylor DE. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990; 94:23–28.

18. Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988; 9:28–32.

19. Park JW, Song JY, Hwang HR, Park HJ, Youn HS, Seo JH, et al. Proteomic analysis of thiol-active proteins of Helicobacter pylori 26695. J Bacteriol Virol. 2012; 42:211–223.

20. Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993; 175:2026–2036.

21. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999; 21:33–37.

22. Olczak AA, Wang G, Maier RJ. Up-expression of NapA and other oxidative stress proteins is a compensatory response to loss of major Helicobacter pylori stress resistance factors. Free Radic Res. 2005; 39:1173–1182.

23. Comtois SL, Gidley MD, Kelly DJ. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediationg resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology. 2003; 149:121–129.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download