I. Introduction
Fascial space abscesses are infectious diseases of the oral and maxillofacial regions that require treatment from an oral and maxillofacial surgeon. Fascial space abscesses in the maxillofacial region are usually caused by dental infections, and the causative pathogens are most often oral commensal bacteria, such as gram-positive (G+) cocci or anaerobic gram-negative (G−) bacilli
1. The infection usually begins as pulpitis or periodontitis before spreading to the surrounding tissues
2.
Abscesses that result from dental infections can progress to fascial space abscesses. Localized abscesses can easily and rapidly be treated by incision and drainage (I&D) followed by antibiotic treatment
3. However, when the abscess does not remain localized and is not treated promptly, the infection can spread to other fascial spaces, including the deep neck space, where it becomes difficult to treat. If the infection progresses in this manner, it can cause symptoms such as tracheal obstruction, septicemia, mediastinitis, and necrotizing fasciitis, and can even be life-threatening in severe cases. Accordingly, early diagnosis of dental infection and appropriate treatment, such as I&D, antibiotic therapy, and elimination of the source of infection are important
45.
In addition to surgical treatments such as I&D, the choice of antibiotics is also crucial when treating fascial space abscesses in the maxillofacial region. Although it is important to obtain information from bacterial cultures and antibiotic sensitivity tests, these tests take generally several days, and it is common to administer empirical antibiotics until the test results are obtained
36. However, indiscriminate widespread use of antibiotics has caused major problems due to the emergence of resistant bacteria
7. Thus, indiscriminate use of antibiotics should be avoided, and it is important to choose a suitable antibiotic based on bacterial cultures and antibiotic sensitivity tests in order to treat and prevent dental infectious diseases
1236. To make this choice, we need a better understanding of the types, characteristics, and antibiotic sensitivity of pathogenic bacteria in dental infectious diseases.
This study investigated the types and antibiotic sensitivity of bacteria cultured from patients with dental infectious disease who underwent drainage culture tests. This information will help surgeons choose appropriate antibiotics for the treatment of dental infectious diseases.
Go to :

II. Materials and Methods
This study included patients who visited the Department of Oral and Maxillofacial Surgery at a National Health Insurance Service (NHIS) Ilsan Hospital between 2007 and 2016 and who had an infectious disease of dental origin that was diagnosed as periodontal abscess, vestibular abscess, canine space abscess, buccal space abscess, infratemporal space abscess, pterygomandibular space abscess, submandibular space abscess, sublingual spaces abscess, or submental space abscess. A total of 1,772 patients who underwent bacterial culture tests were included in the study. Abscesses were diagnosed based on clinical tests and radiography that included a panoramic view, periapical view, and computed tomography with contrast.
The bacterial culture tests were performed using an Amies sterile culture transport swab (Copan Diagnostics, Murrieta, CA, USA). Each specimen was collected from the affected area from drainable pus on the oral mucosa, if available, using the sterile culture swab. When I&D was performed, the specimen was collected from the incision site. First, an incision was made in the infected area to approach the abscess pocket. Next, after confirming pus drainage, the specimen was collected.
The culture swab was sent to the Department of Diagnostic Laboratory Medicine, which performed the bacterial culture and antibiotic sensitivity tests. The cultured bacteria were divided into G+, G−, and anaerobic bacteria, and the strains were listed in the order of prevalence within each group. The number of antibiotic-resistant bacteria was analyzed relative to the total number of bacteria that underwent sensitivity testing.
This retrospective study was approved by the institutional review board of the NHIS Ilsan Hospital (NHIMC 2017-01-036). Because of the retrospective nature of this study, which did not use identifying personal information, the institutional review board of the hospital waived the need to obtain written informed consent from the study subjects.
1. Classification of cultured bacteria
After inspection of the bacterial culture results for each patient, we created a list of detected bacteria. The total number of patients was the number of patients with samples that underwent culture testing. The total number of bacteria was the total number of bacterial strains cultured across all patients. The number of patients and bacteria were analyzed according to bacterial strains. When multiple bacterial strains were cultured from a single patient, all of the cultured bacterial strains were included in the list. Patients who showed no bacteria in the culture tests were excluded from the patient count. The data for all patients and bacteria were tabulated and arranged according to bacterial strain.
2. Analysis of bacterial cultures and antibiotic resistance in
fascial space abscesses in the maxillofacial region
Data were collected for Staphylococcus, Streptococcus, Klebsiella, Enterococcus, and Enterobacter strains. The following antibiotics were tested: penicillins (ampicillin, penicillin G, and oxacillin); macrolides (erythromycin); clindamycin; aminoglycosides (gentamicin); quinolones (ciprofloxacin); tetracycline; and cephalosporins (cefoxitin, cefuroxime, and cefazolin). Data were only included for bacteria that had undergone antibacterial sensitivity tests (antibacterial sensitivity tests were not performed for all bacteria). Statistical analyses were performed in IBM SPSS Statistics (ver. 23; IBM, Armonk, NY, USA). Significance was set at P≤0.05. Difference in age was compared by t-tests.
Go to :

III. Results
This study included 1,772 patients who were diagnosed with maxillofacial abscesses at the Department of Oral and Maxillofacial Surgery of the NHIS Ilsan Hospital from 2007 to 2016. The mean patient age (±standard deviation) was 51.9±20.1 years (range, 3–94 years). The study included 920 male patients (51.9%) with a mean age of 48.1±17.9 years and 852 female patients (48.1%) with a mean age of 55.9±21.5 years. The difference in the mean age was significant (P<0.001); the female were older than the male.
Bacterial cultures were performed on pus samples from the abscesses of the 1,772 patients, and 2,489 bacteria strains were detected, including 2,101 G+ and 388 G− bacteria. Among the G+ bacteria, the anaerobes included 15 strains of
Streptococcus and 6 strains of
Staphylococcus ; we also detected the G+ bacilli
Enterococcus faecalis,
Gemella (
Strep.)
morbillorum,
Gemella haemolysans,
Gemella sp.,
Lactobacillus sp., and
Leuconostoc mesenteroides subsp.
cremoris. The detected aerobes included
Bacillus sp.,
Diphtheroid sp.,
Kocuria (
Micrococcus)
kristinae, and
Micrococcus sp.(
Table 1) In the G+
Streptococcus group, 1,806 (86.0%)
Streptococcus viridans strains were detected, with the alpha-hem. group being the most common (1,690; 80.4%). In addition, 142 (6.8%) G+ bacilli and 47 (2.2%) coagulase-negative staphylococci were detected.
Table 1
Gram-positive bacteria cultured from odontogenic maxillofacial abscesses in 1,772 patients

|
Bacterial strain |
Cultured bacteria |
|
Anaerobes |
Streptococcus agalactiae
|
3 (0.1) |
|
Streptococcus anginosus
|
20 (1.0) |
|
Streptococcus constellatus
|
8 (0.4) |
|
Streptococcus gordonii
|
5 (0.2) |
|
Streptococcus intermedius
|
1 (0.0) |
|
Streptococcus mitis
|
2 (0.1) |
|
Streptococcus parasanguinis
|
13 (0.6) |
|
Streptococcus salivarius
|
40 (1.9) |
|
Streptococcus vestibularis
|
5 (0.2) |
|
Streptococcus viridans, alpha-hem.
|
1,690 (80.4) |
|
Streptococcus, beta-hem. Group C
|
2 (0.1) |
|
Streptococcus, beta-hem. Group F
|
11 (0.5) |
|
Streptococcus, beta-hem. Group G
|
4 (0.2) |
|
Streptococcus pneumoniae
|
1 (0.0) |
|
Streptococcus pyogenes
|
1 (0.0) |
|
Gram-positive bacilli
|
142 (6.8) |
|
Staphylococcus aureus
|
35 (1.7) |
|
Staphylococcus epidermidis
|
26 (1.2) |
|
Staphylococcus, coagulase-negative
|
47 (2.2) |
|
Staphylococcus capitis ss. capitis
|
1 (0.0) |
|
Staphylococcus hominis
|
3 (0.1) |
|
Staphylococcus warneri
|
1 (0.0) |
|
Enterococcus faecalis
|
12 (0.6) |
|
Gemella (Strep.) morbillorum
|
4 (0.2) |
|
Gemella haemolysans
|
5 (0.2) |
|
Gemella sp. |
1 (0.0) |
|
Lactobacillus sp. |
2 (0.1) |
|
Leuconostoc mesenteroides ss. cremoris
|
2 (0.1) |
|
Aerobes |
Bacillus sp. |
1 (0.0) |
|
Diphtheroid sp. |
1 (0.0) |
|
Kocuria (Micrococcus) kristinae
|
7 (0.3) |
|
Micrococcus sp. |
5 (0.2) |
|
Total No. of cultured bacteria |
|
2,101 (100) |

Among the G− bacteria, the anaerobes included 6 strains of
Enterobacter as well as
Aeromonas sp.,
Citrobacter braakii,
Citrobacter freundii,
Escherichia coli,
Klebsiella oxytoca,
K. pneumoniae subsp.
pneumoniae,
Leclercia (
Esch.)
adecarboxylata,
Morganella morganii subsp.
sibonii,
Morganella morganii subsp.
morganii,
Pantoea sp.,
Prevotella (
Bacter.)
melaninogenica, and
Shewanella algae. The detected aerobes included
Acinetobacter baumannii,
A. baumannii (
anitratus),
Acinetobacter sp.,
Pseudomonas aeruginosa,
Stenotrophomonas maltophilia,
Neisseria sp., nonpathogenic
Neisseria, and predominant
Neisseria spp.(
Table 2)
Klebsiella pneumoniae subsp. was the most common G− anaerobe detected (57; 14.7%), while
Neisseria sp. was the most common G− aerobe detected (218; 56.2%).
Table 2
Gram-negative bacteria cultured from odontogenic maxillofacial abscesses in 1,772 patients

|
Bacterial strain |
Cultured bacteria |
|
Anaerobes |
Aeromonas sp. |
1 (0.3) |
|
Citrobacter braakii
|
1 (0.3) |
|
Citrobacter freundii
|
6 (1.5) |
|
Enterobacter aerogenes
|
11 (2.8) |
|
Enterobacter amnigenus
|
1 (0.5) |
|
Enterobacter amnigenus 2
|
1 (0.5) |
|
Enterobacter asburiae
|
1 (0.3) |
|
Enterobacter cloacae
|
30 (7.7) |
|
Enterobacter gergoviae
|
2 (0.5) |
|
Escherichia coli
|
6 (1.5) |
|
Klebsiella oxytoca
|
16 (4.1) |
|
Klebsiella pneumoniae ss. pneumoniae
|
57 (14.7) |
|
Leclercia (Esch.) adecarboxylata
|
1 (0.3) |
|
Morganella morganii ss. sibonii
|
2 (0.5) |
|
Morganella morganii ss. morganii
|
2 (0.5) |
|
Pantoea sp. |
2 (0.5) |
|
Prevotella (Bacter.) melaninogenica
|
1 (0.3) |
|
Shewanella algae
|
1 (0.3) |
|
Aerobes |
Acinetobacter baumannii
|
6 (1.5) |
|
Acinetobacter baumannii (anitratus)
|
5 (1.3) |
|
Acinetobacter sp. |
1 (0.3) |
|
Pseudomonas aeruginosa
|
8 (2.1) |
|
Stenotrophomonas maltophilia
|
2 (0.5) |
|
Neisseria sp. |
218 (56.2) |
|
Neisseria, nonpathogenic
|
1 (0.3) |
|
Neisseria spp. predominant
|
3 (0.8) |
|
Total No. of cultured bacteria |
388 (100) |

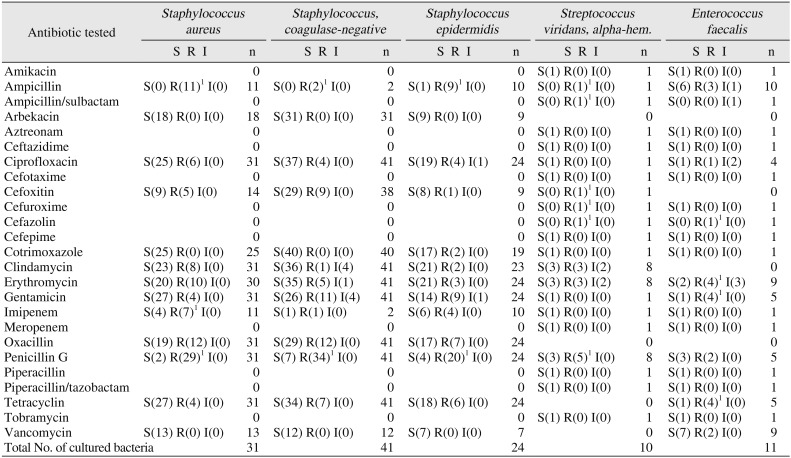
1. Antibiotic sensitivity and resistance of G+ bacteria from maxillofacial abscesses
Antibiotic susceptibility tests of G+ bacteria included tests of 41 coagulase-negative
Staphylococcus strains. All 31 strains tested for susceptibility to arbekacin showed sensitivity, and all 40 strains tested for susceptibility to cotrimoxazole showed sensitivity. Of 41 strains tested for susceptibility to ciprofloxacin, 37 showed sensitivity and 4 showed resistance. In tetracycline susceptibility tests, 34 out of 41 strains showed sensitivity and 7 showed resistance. In cefoxitin susceptibility tests, 29 out of 38 strains showed sensitivity and 9 showed resistance. In clindamycin susceptibility tests, 36 out of 41 strains showed sensitivity, 1 showed resistance, and 4 showed intermediate susceptibility. In erythromycin susceptibility tests, 35 out of 41 strains showed sensitivity, 5 showed resistance, and 1 showed intermediate susceptibility. In gentamicin susceptibility tests, 26 out of 41 strains showed sensitivity, 11 showed resistance, and 4 showed intermediate susceptibility. In oxacillin susceptibility tests, 29 out of 41 strains showed sensitivity and 12 showed resistance. In penicillin G susceptibility tests, 7 out of 41 strains showed sensitivity and 34 showed resistance. In vancomycin susceptibility tests, all 12 strains tested showed sensitivity.(
Table 3)
Table 3
Antibiotic sensitivity and resistance of gram-positive bacteria cultured from odontogenic maxillofacial abscesses in 1,772 patients
|
Antibiotic tested |
Staphylococcus aureus
|
Staphylococcus, coagulase-negative
|
Staphylococcus epidermidis
|
Streptococcus viridans, alpha-hem.
|
Enterococcus faecalis
|
|
S R I |
n |
S R I |
n |
S R I |
n |
S R I |
n |
S R I |
n |
|
Amikacin |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Ampicillin |
S(0) R(11)1 I(0) |
11 |
S(0) R(2)1 I(0) |
2 |
S(1) R(9)1 I(0) |
10 |
S(0) R(1)1 I(0) |
1 |
S(6) R(3) I(1) |
10 |
|
Ampicillin/sulbactam |
|
0 |
|
0 |
|
0 |
S(0) R(1)1 I(0) |
1 |
S(0) R(0) I(1) |
1 |
|
Arbekacin |
S(18) R(0) I(0) |
18 |
S(31) R(0) I(0) |
31 |
S(9) R(0) I(0) |
9 |
|
0 |
|
0 |
|
Aztreonam |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Ceftazidime |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Ciprofloxacin |
S(25) R(6) I(0) |
31 |
S(37) R(4) I(0) |
41 |
S(19) R(4) I(1) |
24 |
S(1) R(0) I(0) |
1 |
S(1) R(1) I(2) |
4 |
|
Cefotaxime |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Cefoxitin |
S(9) R(5) I(0) |
14 |
S(29) R(9) I(0) |
38 |
S(8) R(1) I(0) |
9 |
S(0) R(1)1 I(0) |
1 |
|
0 |
|
Cefuroxime |
|
0 |
|
0 |
|
0 |
S(0) R(1)1 I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Cefazolin |
|
0 |
|
0 |
|
0 |
S(0) R(1)1 I(0) |
1 |
S(0) R(1)1 I(0) |
1 |
|
Cefepime |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Cotrimoxazole |
S(25) R(0) I(0) |
25 |
S(40) R(0) I(0) |
40 |
S(17) R(2) I(0) |
19 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Clindamycin |
S(23) R(8) I(0) |
31 |
S(36) R(1) I(4) |
41 |
S(21) R(2) I(0) |
23 |
S(3) R(3) I(2) |
8 |
|
0 |
|
Erythromycin |
S(20) R(10) I(0) |
30 |
S(35) R(5) I(1) |
41 |
S(21) R(3) I(0) |
24 |
S(3) R(3) I(2) |
8 |
S(2) R(4)1 I(3) |
9 |
|
Gentamicin |
S(27) R(4) I(0) |
31 |
S(26) R(11) I(4) |
41 |
S(14) R(9) I(1) |
24 |
S(1) R(0) I(0) |
1 |
S(1) R(4)1 I(0) |
5 |
|
Imipenem |
S(4) R(7)1 I(0) |
11 |
S(1) R(1) I(0) |
2 |
S(6) R(4) I(0) |
10 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Meropenem |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Oxacillin |
S(19) R(12) I(0) |
31 |
S(29) R(12) I(0) |
41 |
S(17) R(7) I(0) |
24 |
|
0 |
|
0 |
|
Penicillin G |
S(2) R(29)1 I(0) |
31 |
S(7) R(34)1 I(0) |
41 |
S(4) R(20)1 I(0) |
24 |
S(3) R(5)1 I(0) |
8 |
S(3) R(2) I(0) |
5 |
|
Piperacillin |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Piperacillin/tazobactam |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Tetracyclin |
S(27) R(4) I(0) |
31 |
S(34) R(7) I(0) |
41 |
S(18) R(6) I(0) |
24 |
|
0 |
S(1) R(4)1 I(0) |
5 |
|
Tobramycin |
|
0 |
|
0 |
|
0 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
Vancomycin |
S(13) R(0) I(0) |
13 |
S(12) R(0) I(0) |
12 |
S(7) R(0) I(0) |
7 |
|
0 |
S(7) R(2) I(0) |
9 |
|
Total No. of cultured bacteria |
|
31 |
|
41 |
|
24 |
|
10 |
|
11 |

Of the G+ bacteria that were detected, antibiotic susceptibility tests were conducted on 31
Staphylococcus aureus strains. All 18 strains tested for susceptibility to arbekacin showed sensitivity, and all 25 strains tested for susceptibility to cotrimoxazole showed sensitivity. In ciprofloxacin susceptibility tests, 25 out of 31 strains showed sensitivity and 6 showed resistance. In tetracycline susceptibility tests, 27 out of 31 strains showed sensitivity and 4 showed resistance. In cefoxitin susceptibility tests, 9 out of 14 strains showed sensitivity and 5 showed resistance. In clindamycin susceptibility tests, 23 out of 31 strains showed sensitivity and 8 showed resistance. In erythromycin susceptibility tests, 20 out of 30 strains showed sensitivity and 10 showed resistance. In gentamicin susceptibility tests, 27 out of 31 strains showed sensitivity and 4 showed resistance. In oxacillin susceptibility tests, 19 out of 31 strains showed sensitivity and 12 showed resistance. In imipenem susceptibility tests, 4 out of 11 strains showed sensitivity and 7 showed resistance. In vancomycin susceptibility tests, all 13 strains tested showed sensitivity. For penicillin G susceptibility tests, 2 out of 31 strains showed sensitivity and 29 showed resistance. For ampicillin susceptibility tests, all 11 strains tested showed resistance. (
Table 3)
For the G+ bacteria that were detected, antibiotic susceptibility tests were conducted on 24
Staphylococcus epidermidis strains. In arbekacin susceptibility tests, all 9 strains tested showed sensitivity. In cotrimoxazole susceptibility tests, 17 out of 19 strains showed sensitivity and 2 showed resistance. In ciprofloxacin susceptibility tests, 19 out of 24 strains showed sensitivity, 4 showed resistance, and 1 showed intermediate susceptibility. In tetracycline susceptibility tests, 18 out of 24 strains showed sensitivity and 6 showed resistance. In cefoxitin susceptibility tests, 8 out of 9 strains showed sensitivity and 1 showed resistance. In clindamycin susceptibility tests, 21 out of 23 strains showed sensitivity and 2 showed resistance. In erythromycin susceptibility tests, 21 out of 24 strains showed sensitivity and 3 showed resistance. In gentamicin susceptibility tests, 14 out of 24 strains showed sensitivity, 9 showed resistance, and 1 showed intermediate susceptibility. In oxacillin susceptibility tests, 17 out of 24 strains showed sensitivity and 7 showed resistance. In penicillin G susceptibility tests, 4 out of 24 strains showed sensitivity and 20 showed resistance. In imipenem susceptibility tests, 6 out of 10 strains showed sensitivity and 4 showed resistance. In vancomycin susceptibility tests, all 7 strains tested showed sensitivity. In ampicillin susceptibility tests, 9 out of 10 strains showed resistance.(
Table 3) In penicillin G susceptibility tests, 3 out of 8
S. viridans strains showed sensitivity and 5 showed resistance.
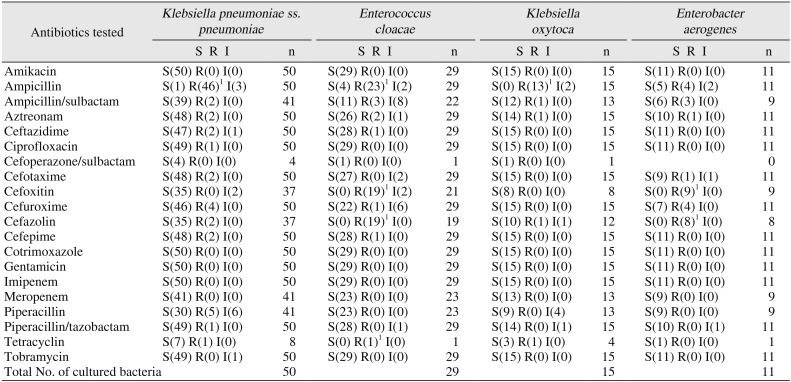
2. Antibiotic sensitivity and resistance of G− bacteria from maxillofacial abscesses
In the antibiotic susceptibility tests on the G− bacteria that were detected, tests were conducted on 50
K. pneumoniae subsp.
pneumoniae strains. All 50 strains tested for amikacin, cotrimoxazole, gentamicin, and imipenem susceptibility showed sensitivity. All 41 strains tested for meropenem susceptibility showed sensitivity. In cefuroxime susceptibility tests, 46 out of 50 strains showed sensitivity and 4 showed resistance. In piperacillin susceptibility tests, 30 out of 41 strains showed sensitivity, 5 showed resistance, and 6 showed intermediate susceptibility. In ampicillin susceptibility tests, 1 out of 50 strains showed sensitivity, 46 showed resistance, and 3 showed intermediate susceptibility. For all other antibiotics tested, the number of strains that showed resistance was less than two.(
Table 4)
Table 4
Antibiotic sensitivity and resistance of gram-negative bacteria cultured from odontogenic maxillofacial abscesses in 1,772 patients
|
Antibiotics tested |
Klebsiella pneumoniae ss. pneumoniae
|
Enterococcus cloacae
|
Klebsiella oxytoca
|
Enterobacter aerogenes
|
|
S R I |
n |
S R I |
n |
S R I |
n |
S R I |
n |
|
Amikacin |
S(50) R(0) I(0) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Ampicillin |
S(1) R(46)1 I(3) |
50 |
S(4) R(23)1 I(2) |
29 |
S(0) R(13)1 I(2) |
15 |
S(5) R(4) I(2) |
11 |
|
Ampicillin/sulbactam |
S(39) R(2) I(0) |
41 |
S(11) R(3) I(8) |
22 |
S(12) R(1) I(0) |
13 |
S(6) R(3) I(0) |
9 |
|
Aztreonam |
S(48) R(2) I(0) |
50 |
S(26) R(2) I(1) |
29 |
S(14) R(1) I(0) |
15 |
S(10) R(1) I(0) |
11 |
|
Ceftazidime |
S(47) R(2) I(1) |
50 |
S(28) R(1) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Ciprofloxacin |
S(49) R(1) I(0) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Cefoperazone/sulbactam |
S(4) R(0) I(0) |
4 |
S(1) R(0) I(0) |
1 |
S(1) R(0) I(0) |
1 |
|
0 |
|
Cefotaxime |
S(48) R(2) I(0) |
50 |
S(27) R(0) I(2) |
29 |
S(15) R(0) I(0) |
15 |
S(9) R(1) I(1) |
11 |
|
Cefoxitin |
S(35) R(0) I(2) |
37 |
S(0) R(19)1 I(2) |
21 |
S(8) R(0) I(0) |
8 |
S(0) R(9)1 I(0) |
9 |
|
Cefuroxime |
S(46) R(4) I(0) |
50 |
S(22) R(1) I(6) |
29 |
S(15) R(0) I(0) |
15 |
S(7) R(4) I(0) |
11 |
|
Cefazolin |
S(35) R(2) I(0) |
37 |
S(0) R(19)1 I(0) |
19 |
S(10) R(1) I(1) |
12 |
S(0) R(8)1 I(0) |
8 |
|
Cefepime |
S(48) R(2) I(0) |
50 |
S(28) R(1) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Cotrimoxazole |
S(50) R(0) I(0) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Gentamicin |
S(50) R(0) I(0) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Imipenem |
S(50) R(0) I(0) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Meropenem |
S(41) R(0) I(0) |
41 |
S(23) R(0) I(0) |
23 |
S(13) R(0) I(0) |
13 |
S(9) R(0) I(0) |
9 |
|
Piperacillin |
S(30) R(5) I(6) |
41 |
S(23) R(0) I(0) |
23 |
S(9) R(0) I(4) |
13 |
S(9) R(0) I(0) |
9 |
|
Piperacillin/tazobactam |
S(49) R(1) I(0) |
50 |
S(28) R(0) I(1) |
29 |
S(14) R(0) I(1) |
15 |
S(10) R(0) I(1) |
11 |
|
Tetracyclin |
S(7) R(1) I(0) |
8 |
S(0) R(1)1 I(0) |
1 |
S(3) R(1) I(0) |
4 |
S(1) R(0) I(0) |
1 |
|
Tobramycin |
S(49) R(0) I(1) |
50 |
S(29) R(0) I(0) |
29 |
S(15) R(0) I(0) |
15 |
S(11) R(0) I(0) |
11 |
|
Total No. of cultured bacteria |
|
50 |
|
29 |
|
15 |
|
11 |

For the G− bacteria that were detected, antibiotic susceptibility tests were conducted on 29
Enterobacter cloacae strains. All 29 strains tested for amikacin, ciprofloxacin, cotrimoxazole, gentamicin, imipenem, and tobramycin susceptibility showed sensitivity. Moreover, all 23 strains tested for meropenem and piperacillin susceptibility showed sensitivity. In ampicillin susceptibility tests, 4 out of 29 strains showed sensitivity, 23 showed resistance, and 2 showed intermediate susceptibility. In ampicillin/sulbactam susceptibility tests, 11 out of 22 strains showed sensitivity, 3 showed resistance, and 8 showed intermediate susceptibility. In cefoxitin susceptibility tests, 19 out of 21 strains showed resistance and 2 showed intermediate susceptibility, with no strains showing sensitivity. In cefazolin susceptibility tests, all 19 strains tested showed resistance. For all other antibiotic susceptibility tests, the number of strains that showed resistance was less than two.(
Table 4)
Go to :

IV. Discussion
Odontogenic infectious diseases in the oral and maxillofacial regions caused by bacteria (mostly oral resident bacteria) are one of the most common diseases seen by dentists
8. Dental infectious diseases can be resolved easily if they can be successfully localized. However, an infection that spreads rapidly can advance to a secondary site and cause airway obstruction, sepsis, mediastinitis, and necrotizing fasciitis
49. Therefore, a timely accurate diagnosis and effective treatment are important.
Treatments for odontogenic infections involve I&D followed by antibiotic therapy, which is a very important aspect of odontogenic infection treatment
5678. Accordingly, the objective of the present study was to investigate the types of bacteria that cause odontogenic infectious diseases and to determine their antibiotic susceptibilities in order to select appropriate antibiotic treatment. The study cohort comprised patients who visited the emergency room or the Department of Oral and Maxillofacial Surgery in the NHIS Ilsan Hospital over a 10-year period (2007 to 2016). It only included patients who had odontogenic infectious diseases in the oral and maxillofacial regions and who underwent I&D with swab culture and bacterial culture tests.
A total of 2,489 bacterial strains were detected in the 1,772 patients, for an average of 1.4 bacterial strains per patient. In general, the most appropriate method for bacterial culture is needle aspiration in the infected area. When sampling bacteria using culture swabs, as in the present study, there is a chance of contamination by normal oral bacteria. Compared to the aspiration method, the swab method detects fewer bacteria per patient. With swab methods, 1 to 1.6 bacterial strains per patient can be detected
3. In the present study, various types of bacteria were detected by culture, including aerobes, anaerobes, and facultative anaerobes. This is consistent with other studies that found a wide spectrum of bacteria in odontogenic infections in the oral and maxillofacial regions, including aerobes, anaerobes, and a mixture of aerobes and anaerobes
4567.
Identification of bacteriological patterns is important for treating odontogenic infectious diseases
8. Numerous studies have shown that
Staphylococcus plays a major role in odontogenic infections, although various other bacteria are also involved
3891011. In the present study,
Streptococcus strains were the most common bacteria found. In another study, a comparison of bacterial culture test results between an odontogenic infection group and a control group found more strict and facultative anaerobes in the odontogenic infection group, especially
S. anginosus and hemolytic streptococci
1. The largest group of bacteria in odontogenic infection patients belonged to the
S. viridans group, similar to the findings in the present study. Notably, the results of antibiotic susceptibility tests were similar between the infected and control groups
1.
In the current study, antibiotic susceptibility tests of G+ bacteria showed that Staphylococci were resistant to penicillin G and ampicillin. In penicillin G susceptibility tests, 7 out of 41 coagulase-negative Staphylococcus strains showed sensitivity and 34 showed resistance. In other penicillin G susceptibility tests, 2 out of 31 S. aureus strains showed sensitivity and 29 showed resistance. In ampicillin susceptibility tests, all 11 S. aureus strains showed resistance. In other ampicillin susceptibility tests, 9 out of 10 S. epidermidis strains showed resistance.
In the present study, when 8 bacterial strains of
S. viridans,
alpha-hem. were tested for clindamycin and erythromycin susceptibility, 3 out of 8 strains showed sensitivity, while 3 showed resistance and 2 showed intermediate susceptibility. In penicillin G susceptibility tests, 3 out of 8 strains showed sensitivity and 5 showed resistance.
S. viridans was previously found to show 77% sensitivity to penicillin and erythromycin and 100% sensitivity to cefepime
12. The number of bacterial strains with antibiotic resistance is gradually increasing
37
When selecting an antibiotic for patients with odontogenic infections, an empirical antibiotic may be used before the bacterial culture test results become available. Since various types of bacteria are involved in odontogenic infections, the use of broad-spectrum empirical antibiotics is recommended. Penicillin has traditionally been considered the first-choice antibiotic for the treatment of odontogenic infections and is still considered the primary choice in emergency cases. In the present study, Staphylococcus and Streptococcus showed resistance to penicillin G and ampicillin. Physicians should consider this result when deciding on antibiotic therapy for odontogenic abscess.
As seen in this study, penicillin is not always effective against anaerobes. Due to recent increases in the number of bacterial strains that are resistant to penicillin, metronidazole or amoxicillin clavulanic acid may be used instead
8. Notably, metronidazole is highly effective against anaerobes in normal flora when treatment of abscess is initiated
12. In cases in which the initial empirical antibiotic administration does not treat the condition, the microbiological profile must be determined, and an antibiotic susceptibility test must be performed. If a bacterial culture test is not performed, switching to another broad-spectrum antibiotic is recommended, such as amoxicillin, clavulanic acid, or clindamycin
812.
Our findings suggest that when treating odontogenic infectious diseases in the oral and maxillofacial region, it is important to choose the right antibiotic based on the results of a mandatory bacterial culture test. However, antibiotic therapy should be administered even before the bacteria culture test results become available. This study showed that a high percentage of bacterial strains are resistant to penicillin G, so it is appropriate to use an antibiotic other than penicillin as the first-choice treatment.
In the antibiotic susceptibility tests on G− bacteria, a high percentage of bacterial strains were resistant to ampicillin. Ampicillin susceptibility tests were conducted on 50 K. pneumoniae subsp. pneumoniae strains: 1 out of 50 strains showed sensitivity, 46 showed resistance, and 3 showed intermediate susceptibility. Ampicillin susceptibility tests were also conducted on 29 E. cloacae strains: 4 showed sensitivity, 23 showed resistance, and 2 showed intermediate susceptibility. For cefoxitin, 19 out of 21 strains showed resistance, 2 showed intermediate susceptibility, and no strains showed sensitivity. For cefazolin, all 19 E. cloacae strains tested showed resistance, with no strains showing sensitivity. Based on these data, we recommend choosing a broad-spectrum antibiotic with low resistance, such as second- and third-generation cephalosporins, clindamycin, or quinolone.
These results may be useful in treatment of infections associated with bisphosphonate-related osteonecrosis of the jaws and osteomyelitis, when the necrotic bone or fistulas to the bone associated with infection, as evidenced by pain and erythema in the region, with or without purulent drainage, are exposed. This result may also be helpful when considering prophylactic antibiotics in oral and maxillofacial surgery, including third molar extraction
13. This study has some limitations. Antibiotic susceptibility tests were not performed on samples from all patients, so we did not determine the antibiotic susceptibility of all bacteria. Our results can be used to reduce the misuse of antibiotics for infections in the maxillofacial region, and additional studies with larger datasets will help confirm and extend our results.
Go to :









 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download