Introduction
Microarray analysis is a high-capacity system to monitor the expression levels of many genes simultaneously. Complementary DNAs on glass are used to quantitatively measure relative expression levels of corresponding genes; microarrays can be used to measure expression levels of thousands of genes in two or more tissue samples [
1]. Although subsequent processes, such as differential degradation of mRNA in the cytoplasm and differential translation, are known to regulate gene expression, considerable knowledge can be gained by estimating relative quantities of mRNA species in populations of cells [
2]. Recent studies have shown that resting and activated microglia show distinct phenotypes in the absence of nervous pathologies [
3]. Activation of microglia by cerebral inflammation results in substantial loss of synaptic contact with adjacent neurons, suggesting a role for microglia in the initiation of synaptic stripping [
4]. In addition, astrocytes exhibit biochemical and morphological changes in response to axotomy [
5].
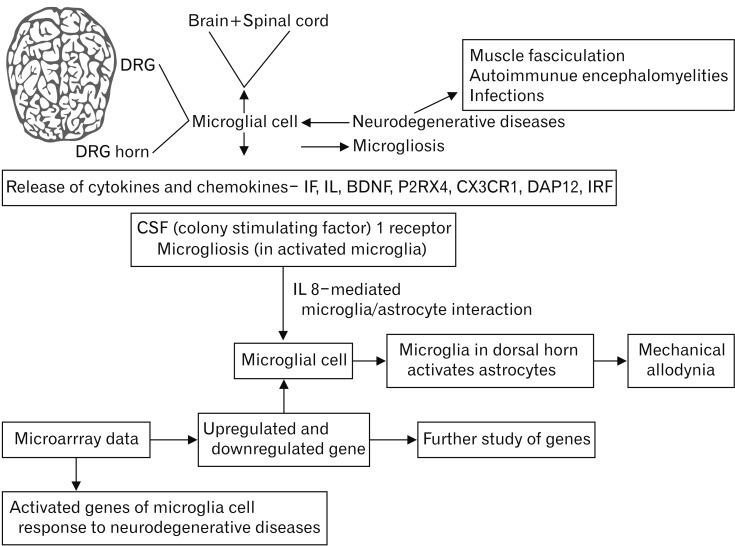
Microglial activation and proliferation in response to neurodegenerative diseases (
Fig. 1) exert stimulatory effects through the dysregulation of chemical signaling using phagocytic products [
6]. Evidence has accumulated regarding the role of microglia in altered synaptic remodeling, connectivity, and network functions, which contribute to the onset of chronic pain-induced neurodegenerative disease; the underlying molecular and cellular mechanisms have become clear increasingly [
7]. In addition, microglia have active roles in vital brain regions that are affected by memory-related aspects of neurodegenerative diseases. A previous overview of gene expression patterns in neurodegenerative diseases using microarray gene expression analysis provided information regarding major classes of transcriptional alterations observed in the cerebral cortex and cerebellum; these alterations were presumed to represent changes that occur throughout the central nervous system during aging [
8]. In the neocortex and cerebellum, aging has been associated with activation of transcriptional responses in ways similar to those observed in human neurodegenerative disorders, such as the induction of lysosomal proteases, chaperones, and inflammatory factors [
9]. The inflammatory response in the central nervous system is mediated by microglial activation and cytokine release, primarily including interferons and interleukins [
10]. In Alzheimer's disease (AD), the responsible genes are calmodulin (
CALM3), calnexin (
CANX), AP-2 complex subunit beta (
AP2B1), and apolipoprotein E (
APOE). These genes have been shown by Cufflinks to exhibit significant splicing between normal and AD tissue in the whole brain, frontal lobes, and temporal lobes [
11]. In spinal motor neurons in amyotrophic lateral sclerosis (ALS), the most upregulated gene is
hsa-mir-338-3p, which showed a 12.7-fold change. This analysis of ALS enables to investigate cellular events that affect cell type-specific gene expression profiles in neurodegenerative disease [
12]. In several research studies, spinal motor neurons and homogenates from ALS patients were analyzed, and cy5-labeled cDNA probes were synthesized from control samples. The data for each expressed gene obtained from microarray analysis were then expressed as the ratios of the values of individual ALS patients [
12].
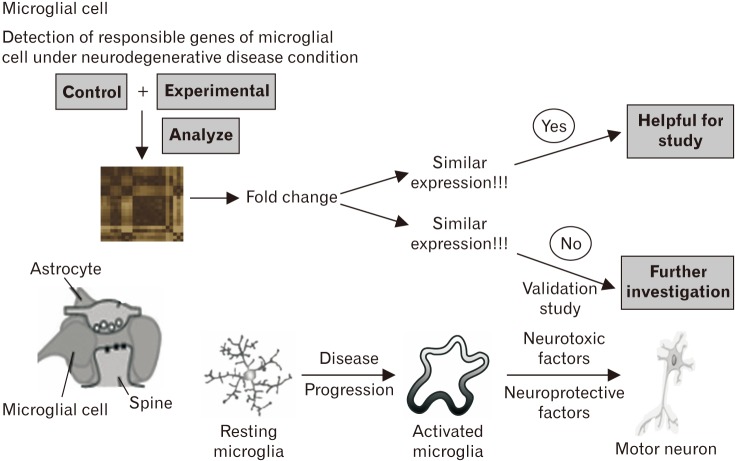 | Fig. 1Microarray study of microglial cell in neurodegenerative disease. This figure describes the activation of microglia is a hallmark of brain pathology. Under neurodegenerative disease condition, the inflammatory response is mediated by the activated microglia. This figure illustrates the gene expression pattern by microarray study in microglial cell under neurodegenerative diseases.
|
After nerve injury-induced neurodegeneration, microglia transform to reactive states through the gene expression of cell-surface receptors and proinflammatory cytokines [
13]. Inhibitory factors and the expression of microglial molecules can strongly suppress a neurodegenerative disorder marked by tactile allodynia to mechanical stimuli. In patients with Sandhoff disease, microglial activation precedes acute neurodegeneration. Microarray gene expression profiling of patients with Sandhoff disease showed that cathepsin, serine protease inhibitor 2-1, metallothionein 1, glycoprotein 49A, and CD68 were highly expressed, with corresponding fold changes of 5.5, 4.9, 4.7, 4.5, and 4.5 [
14]. Microarray gene expression profiling of patients with myoclonic epilepsy (EPMI) showed that
Wfdc17,
lyz2,
C3ar1,
Mpeg1,
ccl3, and
Etnppl were highly expressed, with corresponding fold change of 2.93, 2.28, 1.97, 1.75, and 1.73 [
15].
In a recent gene microarray analysis, abundant transcripts for the proinflammatory cytokine osteopontin (
OPN) were found in plaques of multiple sclerosis (MS) patients' brains [
16].
OPN may participate in MS, as
OPN-deficient mice were resistant to progressive experimental autoimmune encephalomyelitis [
16]. The protein encoded by
OPN is a potent modulator of autoimmune demyelination, and its expression in both neurons and microglia may influence neuronal degeneration in patients who are susceptible to MS [
17].
OPN expression may also affect disease course and severity; increased expression levels of
OPN have been found in the spinal fluid of MS patients during relapses [
18].
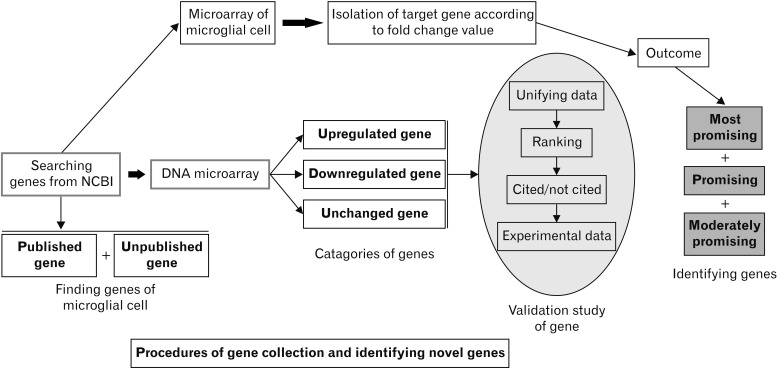
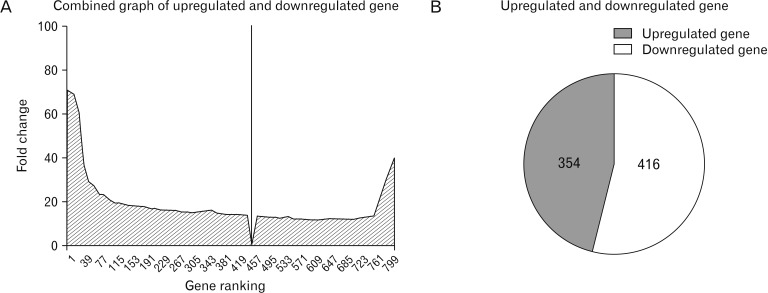
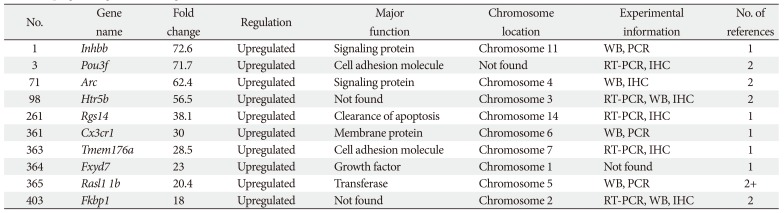
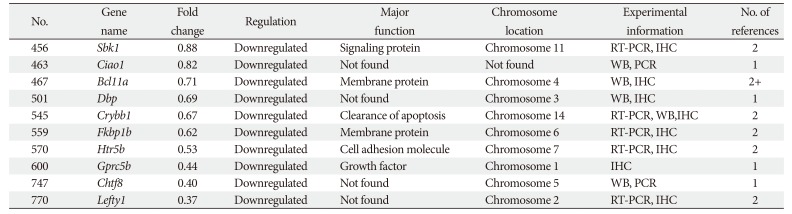
This synthesis of microarray studies of microglial cell gene expression uses in vitro and in vivo sample data from published articles. In those published articles, differential gene expression levels in animal models were compared between control animals and animals exposed to experimental conditions of neuronal damage using microarray techniques; for the current study, fold-change values were acquired from published articles in PubMed and Web Med. In microarray studies, genes are classified according to upregulated and downregulated fold-change values. Here, genes are categorized as upregulated when their fold-change values are >1, and as downregulated when their fold-change values are <1. Genes are categorized as unchanged when their fold change values are equal to 1. Among the 770 total genes examined here, 456 were upregulated and 314 were downregulated.
In this study, published microglial cell gene expression microarray data are analyzed; preliminary microarray data are compared with experimentally validated data from realtime polymerase chain reaction (RT-PCR) or quantitative PCR (qPCR). This comparison does not entirely elucidate the responsible genes or particular characteristics of these genes in each disease. To identify genes of microglial cells that participate in disease, further validation is necessary using Western blotting (WB), immunohistochemistry (IHC), or in situ hybridization (ISH). The principal aim of this study was to identify novel genes and validate their expression levels in microglial cells using randomly selected studies of microarray data. The findings of this study can provide new insights into the origin and phenotypes of microglia in health and in multiple neurodegenerative diseases.
Go to :

Discussion
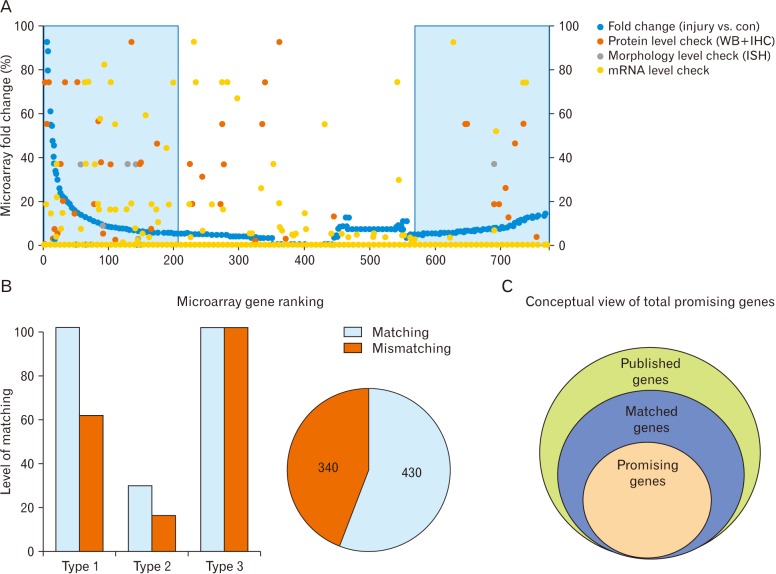
Some microarray data showed expression levels that matched those in experimental studies; these could be useful in further studies of microglial cell function. In the analyses of all 770 published genes, we found that only 15% of all upregulated and downregulated genes were important for use in future microglial cell studies. Mismatched genes in this study were presumed to have no pathological role in disease. Moreover, matched genes were presumed to be useful as target genes in treatment of AD, ALS, MS, and EPMI neurodegenerative diseases. For example, the apolipoprotein gene (APP) was identified as a matched gene. This is consistent with the use of APP as the target gene for AD, and it has indeed been useful in elucidating the pathology of AD.
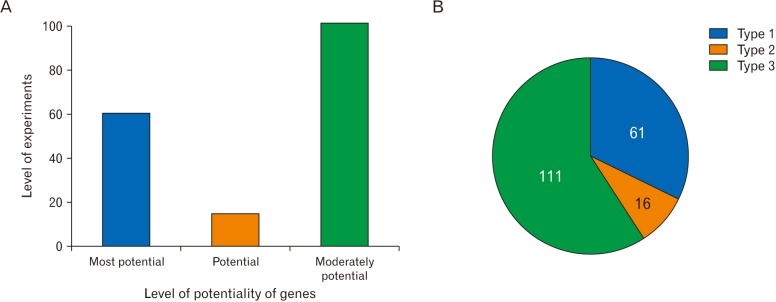
Microarray analysis of microglial genes can identify genes potentially responsible for nerve degeneration. This study included initial categorization of microarray results on microglial cell expression, as well as an arrangement of overall upregulated and downregulated genes. These genes were further categorized based on the extent of experimental studies found for each specific gene. Type 1 genes were those for which protein levels were checked by WB and IHC; type 2 genes were those for which morphological expression levels were checked; type 3 genes were those for which mRNA expression levels were checked. These experimental studies helped to establish the potential of the genes for use in future disease treatment based on the microglial cell response. This big data analysis established classifications of different kinds of genes and their characteristics and functions, such as upregulation or downregulation. In addition, some genes act differently based on experimental conditions, and this study provided further details regarding these genes. These findings may be useful for detection of gene expression in future experimental studies and thus for the proper diagnosis and treatment of diseases.
This study showed that some genes appear robust in microarray studies, but may not exhibit similar expression in experimental studies. Genes with matching data on expression from microarray analysis and experimental studies may be useful in further analyses of microglial cell function. Although no experimental methods were used in this study, the aggregated gene expression data in this study may enable further analyses of the microglial cell response to neurodegeneration.
Go to :






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download