Abbreviations
ADCC
CDC
CDR
CSC
CTX
EGFR
Fc
HCC
HER2
HMGB1
i.p.
i.v.
ICB
L
MTS
NSCLC
PBST
POSTECH
TME
W
INTRODUCTION
MATERIALS AND METHODS
Mice
Cell lines and the generation of Kiatomab and its isotype variant
Epitope determination and sequencing variable regions in Kiatomab
Tumor models and treatments
Flow cytometry
RESULTS AND DISCUSSION
Identification and characterization of the linear epitope and variable regions of Kiatomab
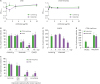
Figure 1
Characterization of Kiatomab and KIAA1114 expression in mouse cancer cells. (A) Amino acid sequences of the extracellular domain of human KIAA1114 and corresponding mouse KIAA1114 sequences were aligned. Among their overlapping sequences, possible linear epitope regions were determined by surface accessibility prediction (dotted lines) and Bepipred linear epitope prediction (solid lines) programs. Subsegments simultaneously identified by two prediction tools were synthesized as peptides (boxes) and subjected to direct ELISA using Kiatomab. (B) The binding of Kiatomab at 3µg/ml to various peptides and (C) dose-dependent Kiatomab binding are shown. (D) The variable regions of Kiatomab were clarified by RT-PCR with mRNA from the hybridoma, and CDR was determined through the Kabat sequence database. (E) KIAA1114 expression in various murine cancer cell lines was determined by Kiatomab. Representative data are shown from two or three independent experiments.

Kiatomab inhibits metastasis of KIAA1114-overexpressing cancer cells through the host immune system
Figure 2
Immune-mediated suppression of tumor growth by Kiatomab in pulmonary metastasis models. (A) Different doses of Kiatomab or control Ig were co-cultured with tumor cells for 72 h. The viability of cells was determined by MTS assay. (B-E) 2×105 tumor cells were i.v. injected into (B) BALB/c, (C) C57BL/6, and (D, E) NOD/SCID mice. One day later, the indicated doses of Kiatomab (10 µg for NOD/SCID mice), its F(ab′)2 fragment, or control Ig were i.p. injected into tumor-bearing mice. On day 14 after tumor injection, lungs were extracted and the number of metastatic nodules on the lung surface was counted (n=7). (E) Tumor-bearing mice were treated with 10 µg of Kiatomab and control Ig at the indicated time points.
Combination with Kiatomab and CTX enhances the antitumor efficacy in both metastasis and solid cancer models
Figure 3
The combination of Kiatomab with CTX improves antitumor activity in metastatic tumor models. (A-C) 2×105 CT26-HER2/neu or (D) B16F10 tumor cells were i.v. injected into BALB/c or C57BL/6 mice. Tumor-bearing mice were i.p. treated with 10 µg Kiatomab at day 0 or day 3, 30 mg/kg CTX at day 4, or both agents at the designated time points. Control group was injected with control Ig or PBS for CTX. Mice were either sacrificed (B, D) at day 14 for metastatic nodule counting (n=7) or (C) maintained for 10 wk for survival monitoring (n=10).

Figure 4
CTX combination and isotype switching augment the antitumor activity of Kiatomab in a solid tumor model; 5×105 CT26 cells were subcutaneously injected into BALB/c mice. (A) On day 4 and 11 after tumor inoculation, mice were i.p. treated with 30 µg Kiatomab, 30 mg/kg CTX, or a combination of 2 agents. Tumor growth was monitored until the mean of PBS-treated group reached 1,000 mm3 (day 18) (n=7). (B) On day 4 and 11, mice were injected with 30 µg of Kiatomab, 10 or 30 µg of IgG2a isotype switch variant. Mice were maintained up to 18 days for tumor growth measurement (n=7).
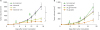




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download