Abstract
We report a case of cellulitis caused by a novel Cupriavidus species identified using whole-genome sequence analysis. Subcutaneous tissue biopsies from the left lower leg of a 67-year-old man who suffered from cellulitis were cultured. Round, convex, gray and non-hemolytic colonies were recovered after 72-h incubation. 16S rRNA sequence analysis showed 98.6% similarity with Cupriavidus basilensis DSM 11853(T) in the NCBI database and 99.9% similarity with C. basilensis KF708 in the EzBioCloud database. Genomic analysis using the MiSeq platform (Illumina, USA) and the TrueBac ID database (ChunLab, Korea) revealed that the average nucleotide identity (ANI) of this strain with C. basilensis DSM 11853(T) was 87.6%. The patient was treated with oral cefditoren pivoxil for 9 weeks. This study is the first to report cellulitis caused by Cupriavidus species strain J1218.
Cupriavidus is a genus of aerobic gram-negative glucose-nonfermenting bacillus common in the environment but rarely isolated from clinical specimens. Clinical manifestations of infection with Cupriavidus species are varied and include pneumonia, bacteremia, meningitis, and septicemia in children [123]. Muscular abscess in a renal transplant recipient and airway infection with a Cupriavidus species in patients with cystic fibrosis suggest that immunodeficiency may be a prior condition for infection by this genus [34567].
We describe the first case of class II cellulitis in an immunocompetent patient caused by a novel Cupriavidus species identified using whole-genome sequencing.
A 67-year-old man complained of fever, redness, swelling, and pain in his left lower leg for 2 weeks; the lesion did not respond to antibiotic therapy. He had no underlying disease other than benign prostatic hyperplasia. He was often exposed to freshwater during fishing and gardening. Erythema remained after 10 days of intravenous and oral ceftriaxone treatment. Intravenous and oral levofloxacin treatment was then given for 6 weeks, but the lesion persisted. A magnetic resonance imaging (MRI) scan with enhancement of the lower leg revealed prominent swelling of the cutaneous and subcutaneous layer and non-enhancing fluid in the mid- to distal calf. Tissue was obtained from the lower leg via ultrasound-guided biopsy for bacterial and acid-fast bacterial culture. Round, convex, gray, non-hemolytic colonies were visible on BAP agar after 72 h incubation at 35℃ (Fig. 1). Vitek MS IVD v3.0 (bioMérieux, Manchester, UK) failed to identify these colonies, while the Vitek 2 system (bioMérieux, Hazelwood, MO, USA) identified them as Cupriavidus pauculus. For 16S rRNA sequencing, genomic DNA was extracted using a Genedia Mycobacteria DNA Prep kit (Green Cross Medical Science co., Eumseong, Korea) and amplification of the 16S rRNA gene was performed using the following universal primer sets: 4F: 5′-TTGGAGAGTTT GATCCTGGCTC-3′, 534R: 5′-TACCGCGGCTGCTGGCAC-3′, 27F: 5′-AGAGTTTGATCMTGGCTCAG-3′ and 801R: 5′-GGC GTGGACTTCCAGGGTATCT-3′. The resulting sequences were compared with the 16S rRNA sequences (713 bp) of related taxa, which were obtained from the GenBank database and EzBioCloud database. Sequences showed 99.9% similarity with C. basilensis strain KF708 (GenBank accession no. AB109778) in EzBioCloud and 98.6% similarity with C. basilensis strain DSM 11853T (GenBank accession no.: NR_025138) in the National Center for Biotechnology Information (NCBI) database. A neighbor-joining phylogenetic tree was constructed using BLAST pairwise alignments (Fig. 2).
A paired-end library (insert size: 300 bp) was constructed from genomic DNA using a TruSeq DNA LT Sample Preparation kit (Illumina, San Diego, CA, USA). The nucleotide sequences of this strain were determined by synthesis on the MiSeq platform (Illumina). Gene discovery and functional annotation pipeline of the whole-genome assembly were performed using data from the EzBioCloud database. For average nucleotide identity (ANI) calculations, the query genome was cut into small fragments (1,020 bp), and the BLAST algorithm was used to select the highest-scoring pair between two genomic sequences. The size of the genome was 7,960,659 bp, with 795 contigs, and the GC content of the coding sequences was 68.6%.
The first taxon identified by TrueBac ID was Cupriavidus genomospecies BBQM_s (KF708), according to the overall genome relatedness index (OGRI). The ANI of Cupriavidus strain J1218 with Cupriavidus genomosp. BBQM was 98.6%, while it shared 87.6% ANI with C. basilensis DSM 11853T, according to genome analysis using the TrueBac ID database (ChunLab, Seoul, South Korea).
The antimicrobial susceptibility test (AST) results of Cupriavidus strain J1218 performed using MicroScan (Beckman Coulter, Brea, CA, USA) are shown in Table 1. The strain was suspected to be resistant to ciprofloxacin because the patient's lesion persisted despite continued treatment with ceftriaxone for 10 days and levofloxacin for 6 weeks. However, an AST of Cupriavidus strain J1218 performed using MicroScan showed susceptibility to all antimicrobial agents, including ciprofloxacin, making the choice of treatment difficult. We performed a second AST of strain J1218 using a Sensititre DKMGN panel (Trek Diagnostic Systems; Thermo Fisher Scientific, Inc., East Grinstead, UK) to yield AST data for up to 72 h, taking into account the slow growth characteristics of this strain. However, the minimum inhibitory concentration (MIC) of ciprofloxacin was still less than 0.06 µg/mL (Table 1). Finally, the patient was successfully treated with a 9-week course of oral cefditoren pivoxil.
The criteria for prokaryotic species circumscription are traditionally defined as a DNA-DNA hybridization (DDH) value of at least 70% similarity, and a temperature within 5℃ of the thermal denaturation midpoint [8]. Recently, 16S rRNA sequence similarity of less than 98.7%, and ANI values of 95-96% or a digital DDH of less than 70%, were proposed as criteria for classifying new species based on whole-genome sequencing data [910]. A recent study found that 12% of bacteria prospectively collected from intensive care units in a tertiary care hospital during 1 year were novel genomospecies, according to pairwise ANI analysis using BLAST [11].
In this study, we were unable identify this isolate using Vitek MS despite repeated tests. The Vitek 2 system identified this isolate as C. pauculus, which was included in the Vitek MS v3.0 library. We performed 16S rRNA sequencing analysis to determine the reason for the discrepancy between the Vitek 2 system and Vitek MS. Cupriavidus species are known to be difficult to identify by biochemical tests, making molecular assays necessary for accurate identification of this genus [4].
The 16S rRNA sequencing result was analyzed according to the Clinical and Laboratory Standards Institute (CLSI) guideline MM18-ED2 [12]. This strain was identified as C. basilensis strain KF708 with 99.9% similarity, and could be distinguished from C. basilensis strain DSM 11853 showing 98.9% similarity in the EzBioCloud. In addition, species identification based on the NCBI GenBank database limiting to sequences from type material of this strain showed a similarity of 98.6% with C. basilensis DSM 11853T. The phylogenetic tree created by NCBI GenBank revealed it to be an unknown strain.
A whole genome analysis classed the sequences as Cupriavidus genomospecies BBQM (KF708) with 98.5% similarity. A previous reclassification study of Cupriavidus suggested that the standard ANI cut-off value of Cupriavidus spp. should be 90% [13]. In the same study, C. basilensis strain KF708 was reclassified as Cupriavidus sp. by a combination of phylogenetic analyses and whole-genome sequence analyses [13]. Genomic analysis revealed that the strain investigated in the current study was a novel Cupriavidus species.
Whole-genome sequencing data of this strain showed 100% identity of carA, ErmE, Brucella suis mprF and OXA-63 genes. Also, it had an adeF gene with 75–76% similarity and the gene was associated with fluoroquinolone resistance with efflux pumps. The AST results for Cupriavidus strain J1218 showed that it is susceptible to all antibiotic agents according to the CLSI guideline M100 [14]. Since differences can exist between genetic and phenotypic data, further study is needed on the relationship between the AST and whole-genome sequencing data.
Patients infected with Cupriavidus species are usually immunocompromised [2357]. Cases of infection with this genus have also been reported in elderly patients without obvious immunodeficiency [1516]. The patient in the present study was immunocompetent, but had experienced frequent injuries to the lower limbs. The patient underwent sustained antimicrobial treatment and the lesion was completely healed after 6 months.
In conclusion, we identified a novel Cupriavidus sp. strain, J1218, using 16S rRNA-based molecular analyses and whole-genome sequencing. This is the first reported case of cellulitis caused by a novel Cupriavidus sp. strain J1218 requiring long-term antibiotic therapy in an immunocompetent patient.
Figures and Tables
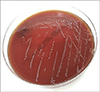 | Fig. 1Circular, convex, gray non-hemolytic colonies of Cupriavidus sp. strain J1218 were grown on BAP after 72 h incubation at 35℃. |
References
1. Duggal S, Gur R, Nayar R, Rongpharpi SR, Jain D, Gupta RK. Cupriavidus pauculus (Ralstonia paucula) concomitant meningitis and septicemia in a neonate: first case report from India. Indian J Med Microbiol. 2013; 31:405–409.
2. D'Inzeo T, Santangelo R, Fiori B, De Angelis G, Conte V, Giaquinto A, et al. Catheter-related bacteremia by Cupriavidus metallidurans. Diagn Microbiol Infect Dis. 2015; 81:9–12.
3. Almasy E, Szederjesi J, Rad P, Georgescu A. A fatal case of community acquired Cupriavidus pauculus pneumonia. J Crit Care Med (Targu Mures). 2016; 2:201–204.
4. Kalka-Moll WM, LiPuma JJ, Accurso FJ, Plum G, van Koningsbruggen S, Vandamme P. Airway infection with a novel Cupriavidus species in persons with cystic fibrosis. J Clin Microbiol. 2009; 47:3026–3028.
5. Karafin M, Romagnoli M, Fink DL, Howard T, Rau R, Milstone AM, et al. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J Clin Microbiol. 2010; 48:1005–1007.
6. Yahya R, Alyousef W, Omara A, Alamoudi S, Alshami A, Abdalhamid B. First case of pneumonia caused by Cupriavidus pauculus in an infant in the Gulf Cooperation Council. J Infect Dev Ctries. 2017; 11:196–198.
7. Tena D, Losa C, Medina MJ, Sáez-Nieto JA. Muscular abscess caused by Cupriavidus gilardii in a renal transplant recipient. Diagn Microbiol Infect Dis. 2014; 79:108–110.
8. Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001; 25:39–67.
9. Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018; 68:461–466.
10. Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009; 106:19126–19131.
11. Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, et al. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet. 2015; 11:e1005413.
12. CLSI. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. CLSI document MM18. Wayne, PA: Clinical and Laboratory Standards Institute;2018.
13. Moriuchi R, Dohra H, Kanesaki Y, Ogawa N. Complete genome sequence of 3-chlorobenzoate-degrading bacterium Cupriavidus necator NH9 and reclassification of the strains of the genera Cupriavidus and Ralstonia based on phylogenetic and whole-genome sequence analyses. Front Microbiol. 2019; 10:133.
14. CLSI. Performance standards for antimicrobial susceptibility testing. CLSI document M100. Wayne, PA: Clinical and Laboratory Standards Institute;2018.
15. Zhang Z, Deng W, Wang S, Xu L, Yan L, Liao P. First case report of infection caused by Cupriavidus gilardii in a nonimmunocompromised Chinese patient. IDCases. 2017; 10:127–129.
16. Kobayashi T, Nakamura I, Fujita H, Tsukimori A, Sato A, Fukushima S, et al. First case report of infection due to Cupriavidus gilardii in a patient without immunodeficiency: a case report. BMC Infect Dis. 2016; 16:493.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download