Abstract
The human respiratory tract hosts both pathogenic and commensal bacteria. The development of well-conserved 16S rRNA sequencing and culture-independent techniques has enabled many achievements in the study of the human microbiome. Microbial composition of the respiratory tract in early childhood has been shown to correlate to respiratory health in later stages of life. This review highlights current understandings of respiratory microbiota development in healthy children, examples of microbial interactions, impacts on the host immune system, and the relationship between respiratory tract microbiome and respiratory health.
Figures and Tables
Fig. 2
Gut-lung communication. The oropharynx, bloodstream, and nerve may serve as routes of immunologic communication between the gut and the lung.
Abbreviations: SCFAs, short-chain fatty acids.
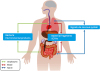
Table 1

References
1. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977; 31:107–133.

2. Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977; 74:5088–5090.

3. Stahl DA, Lane DJ, Olsen GJ, Pace NR. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984; 224:409–411.

4. Woese CR, Olsen GJ. Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol. 1986; 7:161–177.

5. Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990; 345:60–63.

6. Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991; 173:4371–4378.

7. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921.
8. NIH HMP Working Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome Res. 2009; 19:2317–2323.

9. Rogers GB, Shaw D, Marsh RL, Carroll MP, Serisier DJ, Bruce KD. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax. 2015; 70:74–81.

10. Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med. 2013; 187:1382–1387.

11. Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001; 9:206–208.

12. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005; 308:1635–1638.

13. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007; 5:e177.

15. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014; 6:237ra65.

16. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016; 6:23129.

17. Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016; 4:29.

18. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016; 351:1296–1302.

19. Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. 2016; 165:827–841.

20. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010; 107:11971–11975.

21. Bosch AATM, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine. 2016; 9:336–345.

22. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014; 190:1283–1292.

23. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015; 17:704–715.

24. Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012; 129:950–960.

25. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006; 118:511–521.

26. Prevaes SM, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper-Stranders GA, Wyllie AL, et al. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med. 2016; 193:504–515.

27. Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012; 78:6262–6270.

28. Leibovitz E, Greenberg D, Piglansky L, Raiz S, Porat N, Press J, et al. Recurrent acute otitis media occurring within one month from completion of antibiotic therapy: relationship to the original pathogen. Pediatr Infect Dis J. 2003; 22:209–216.

29. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011; 6:e17035.

30. Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004; 363:1871–1872.

31. Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol. 2015; 135:905–12.e11.

32. Spijkerman J, Prevaes SM, van Gils EJ, Veenhoven RH, Bruin JP, Bogaert D, et al. Long-term effects of pneumococcal conjugate vaccine on nasopharyngeal carriage of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis
. PLoS One. 2012; 7:e39730.

33. Greenberg D, Givon-Lavi N, Broides A, Blancovich I, Peled N, Dagan R. The contribution of smoking and exposure to tobacco smoke to Streptococcus pneumoniae and Haemophilus influenzae carriage in children and their mothers. Clin Infect Dis. 2006; 42:897–903.

34. Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al.
Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015; 1:e1400216.
35. Lim MY, Yoon HS, Rho M, Sung J, Song YM, Lee K, et al. Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci Rep. 2016; 6:23745.

36. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012; 486:222–227.

37. Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 2015; 9:1246–1259.

38. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010; 5:e9836.

39. Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010; 5:e15216.

40. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013; 187:1067–1075.

41. Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016; 4:37.

42. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015; 6:e00037-15.

43. Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013; 188:1246–1252.

44. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007; 357:1487–1495.

45. Lohmann P, Luna RA, Hollister EB, Devaraj S, Mistretta TA, Welty SE, et al. The airway microbiome of intubated premature infants: characteristics and changes that predict the development of bronchopulmonary dysplasia. Pediatr Res. 2014; 76:294–301.

46. Payne MS, Goss KC, Connett GJ, Kollamparambil T, Legg JP, Thwaites R, et al. Molecular microbiological characterization of preterm neonates at risk of bronchopulmonary dysplasia. Pediatr Res. 2010; 67:412–418.

47. Mourani PM, Harris JK, Sontag MK, Robertson CE, Abman SH. Molecular identification of bacteria in tracheal aspirate fluid from mechanically ventilated preterm infants. PLoS One. 2011; 6:e25959.

48. Huitzil S, Sandoval-Motta S, Frank A, Aldana M. Modeling the role of the microbiome in evolution. Front Physiol. 2018; 9:1836.

49. Mashima I, Nakazawa F. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe. 2014; 28:54–61.

50. Cook LC, LaSarre B, Federle MJ. Interspecies communication among commensal and pathogenic streptococci. MBio. 2013; 4:e00382-13.

51. Armbruster CE, Hong W, Pang B, Weimer KE, Juneau RA, Turner J, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio. 2010; 1:e00102-10.

52. Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013; 14:631–640.

53. Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP.
Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol. 2016; 7:1230.
54. Deasy AM, Guccione E, Dale AP, Andrews N, Evans CM, Bennett JS, et al. Nasal inoculation of the commensal Neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: a controlled human infection study. Clin Infect Dis. 2015; 60:1512–1520.

55. Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al.
Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010; 465:346–349.

56. Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016; 535:511–516.

57. Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP.
Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. 2016; 7:e01725-15.

58. van den Broek MF, De Boeck I, Kiekens F, Boudewyns A, Vanderveken OM, Lebeer S. Translating recent microbiome insights in otitis media into probiotic strategies. Clin Microbiol Rev. 2019; 32:e00010-18.

59. Sajjan US, Jia Y, Newcomb DC, Bentley JK, Lukacs NW, LiPuma JJ, et al.
H. influenzae potentiates airway epithelial cell responses to rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J. 2006; 20:2121–2123.

60. Blanken MO, Rovers MM, Molenaar JM, Winkler-Seinstra PL, Meijer A, Kimpen JL, et al. Respiratory syncytial virus and recurrent wheeze in healthy preterm infants. N Engl J Med. 2013; 368:1791–1799.

61. Zomer-Kooijker K, van der Ent CK, Ermers MJ, Uiterwaal CS, Rovers MM, Bont LJ, et al. Increased risk of wheeze and decreased lung function after respiratory syncytial virus infection. PLoS One. 2014; 9:e87162.

62. Ni K, Li S, Xia Q, Zang N, Deng Y, Xie X, et al. Pharyngeal microflora disruption by antibiotics promotes airway hyperresponsiveness after respiratory syncytial virus infection. PLoS One. 2012; 7:e41104.

63. Boase S, Jervis-Bardy J, Cleland E, Pant H, Tan L, Wormald PJ. Bacterial-induced epithelial damage promotes fungal biofilm formation in a sheep model of sinusitis. Int Forum Allergy Rhinol. 2013; 3:341–348.

64. Briard B, Heddergott C, Latgé JP. Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus
. MBio. 2016; 7:e00219-16.

65. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015; 12:821–830.

66. Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007; 44:3100–3111.

67. Wang J, Li F, Sun R, Gao X, Wei H, Li LJ, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013; 4:2106.

68. Rice TA, Brenner TA, Percopo CM, Ma M, Keicher JD, Domachowske JB, et al. Signaling via pattern recognition receptors NOD2 and TLR2 contributes to immunomodulatory control of lethal pneumovirus infection. Antiviral Res. 2016; 132:131–140.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download