Abstract
There is an urgent need for accurate and rapid diagnostic assays capable of identifying carbapenemase-producing Enterobacteriaceae (CPE). We assessed the performance of the RESIST-4 O.K.N.V. (OKNV) assay (Coris BioConcept, Gembloux, Belgium) for the identification of oxacillinase (OXA)-48-like-, Klebsiella pneumoniae carbapenemase (KPC)-, New Delhi metallo-β-lactamase (NDM)-, and Verona integron-encoded metallo-β-lactamase (VIM)-producing Enterobacteriaceae grown on sheep blood agar (SBA) and the CHROMagar KPC medium. Sixty-five carbapenem-resistant Enterobacteriaceae (CRE) isolates with characterized carbapenemase content were used to evaluate the OKNV assay. The assay correctly identified all 30 isolates that produced one of the four targeted carbapenemase families. Additionally, it correctly identified 15 isolates that co-produced KPC and NDM, VIM and NDM or OXA-48-like and NDM, but failed to identify an NDM-1 and OXA-232 co-producing Klebsiella pneumoniae isolate. All 16 non-carbapenemase-producing CRE and four CPE isolates exhibited negative results, and no cross-reaction was observed. Overall, the sensitivity and specificity of the assay were 97.8% and 100%, respectively. The OKNV assay is an accurate and rapid assay for identifying OXA-48-like, KPC, NDM, and VIM carbapenemases produced by Enterobacteriaceae isolates cultured on both SBA and the CHROMagar KPC media in the clinical microbiology laboratory.
The global dissemination of carbapenemase-producing Enterobacteriaceae (CPE) is a major public health concern. Accurate and rapid detection of CPE is essential for patient management and for implementation of appropriate infection control measures [12]. Globally, Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), Verona integron-encoded metallo-β-lactamase (VIM), imipenem-resistant Pseudomonas (IMP), and oxacillinase (OXA)-48-like are the most prevalent carbapenemases in Enterobacteriaceae. Thus, it is essential not only to identify carbapenemase production but also to characterize the enzyme. As the identification of CPE in clinical microbiological laboratories based solely on the resistance phenotype is difficult [3], a number of rapid colorimetric assays have been developed for the identification of carbapenemase activity in cultured bacteria. However, a major limitation of this approach is the absence of reliable assays for the identification of OXA-48-like producers [45].
Recently, lateral flow immunochromatographic assays based on monoclonal antibodies generated by immunization of mice have been developed for rapid and easy identification of OXA-48-like, KPC, and NDM carbapenemases. This technology exhibited excellent accuracy (100% sensitivity and 100% specificity) in the identification OXA-48-like, KPC, and NDM producers directly from bacterial colonies within 15 minutes [67]. We evaluated the new RESIST-4 O.K.N.V. (OKNV) multiplex lateral flow assay developed by Coris BioConcept (Gembloux, Gembloux, Belgium), which identifies specific antibodies against OXA-48, KPC, NDM, and VIM carbapenemases directly from different culture media such as sheep blood agar (SBA) and a chromogenic medium. Different types of chromogenic media, such as CHROMagar KPC, chromID CARBA SMART (BioMérieux, Marcy-I'Etoile, France), and Brilliance CRE (ThermoFisher Scientific, Illkirch, France), are often used in rectal surveillance culture for identifying carbapenem-resistant Enterobacteriaceae (CRE) and/or CPE [89].
Globally, the OKNV assay shows 97–100% sensitivity and 100% specificity, with unequivocal results [101112]. Therefore, additional confirmatory assays were not required. Unlike other studies, the number of carbapenemase co-producing isolates included was relatively high, and this is the first study to include isolates grown on a chromogenic medium.
In total, 65 CRE clinical isolates were included in the study (49 CPE and 16 non-CP-CRE). The CRE isolates were obtained from six Hallym University Medical Centers (two hospitals in Seoul, two hospitals in Gyunggi, and one hospital in Gangwon) in Korea between 2012 and 2018, and all isolates were sent to one institution (Kangnam Sacred Heart Hospital, Seoul) and frozen before study. The study protocol was approved by the Institutional Review Board of each institution, which waived the need for informed consent. All isolates were tested for carbapenemase by PCR and DNA sequencing according to previously described methods [1314]. The CPE isolates included nine KPC, eight NDM-1, seven OXA-48-like, five VIM, two IMP, and two Guiana extended-spectrum β-lactamase (GES)-5,carbapenemase variants. In addition, 16 Enterobacteriaceae isolates co-producing carbapenemases (OXA-48-like and NDM [N=9], KPC and NDM [N=5], and VIM and NDM [N=2]) were included. Sixteen non-CP-CRE isolates were also included (Table 1).
The OKNV assay is a multiplex lateral flow immunochromatographic assay for the detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in two lateral flow cassettes (one for OXA-48-like and KPC and the other one for NDM and VIM). The assayed isolates were grown on SBA (SPL Life Sciences, Gyeonggi-do, Korea) and CHROMagar KPC medium (CHROMagar, Paris, France) for 16–24 hours at 37℃, and the assays were performed according to the manufacturer's instructions. An example of the identification results for OXA-48, KPC, NDM, and VIM using the OKNV assay is shown in Fig. 1. The sensitivity and specificity of the assay were calculated for all isolates. Sensitivity was calculated from the number of true-positive isolates, whereas specificity was calculated from the number of true-negative isolates.
All isolates that grew on both SBA and CHROMagar KPC media showed the same results in the OKNV assay. The OKNV assay could identify all isolates that produced one of the four targeted carbapenemase families, including eight OXA-48-like (OXA-48 [N=1], OXA-181 [N=3], and OXA-232 [N=4]), nine KPCs (KPC-2 [N=7] and KPC-4 [N=2]), eight NDMs (all NDM-1), and five VIMs (VIM-1 [N=2], and VIM-2 [N=3]). Additionally, it correctly identified 15 isolates that co-produced KPC and NDM (N=5), VIM and NDM (N=2), or OXA-48-like and NDM (N=8), but failed to identify one K. pneumoniae isolate co-producing NDM-1 and OXA-232 (negative for NDM and positive for OXA-48). One NDM-1-producing Serratia marcescens isolate, one NDM-1 and OXA-232 co-producing Enterobacter cloacae isolate, and one NDM-1 and OXA-232 co-producing K. pneumoniae isolate showed a weak positive NDM band, and two NDM-1 and VIM-2 co-producing E. cloacae isolates showed a weak positive VIM band. All 16 non-CP-CRE and four CPE (two GES-5, one IMP-1, and one IMP-4) were negative for carbapenemase production, and no cross-reaction was observed (Table 1). Based on the overall results, the sensitivity and specificity of the OKNV assay for identifying isolates grown on both SBA and CHROMagar KPC were 97.8% (95% CI, 88.2%–99.9%) and 100% (95% CI, 83.2%–100%), respectively.
The OKNV assay performed well, when using colonies grown on both SBA and CHROMagar KPC. It is possible that the negative result obtained for the NDM-1 and OXA-232 co-producing K. pneumoniae isolate was due to plasmid loss during serial passage [15]. However, subsequent PCR and sequencing analysis of colonies grown on the same culture medium demonstrated that the isolate was NDM-1- and OXA-232-positive. Thus, we confirmed that the NDM-negative result obtained using the OKNV assay was a real false-negative result.
In Korea, KPC, NDM, and OXA-48-like genes are expressed by 71%, 14%, and 9% of CPE, respectively, while VIM and IMP genes are very rare (<2% of CPE) [16]. As the OKNV assay does not identify certain carbapenemases (mostly those belonging to the IMP family), the use of other diagnostic methods, such as PCR, is still required. Furthermore, another lateral flow assay, including IMP-type monoclonal antibodies, is currently under development [17]. Although a few NDM-positive or VIM-positive isolates showed weak positive results, the OKNV assay was easy to use and provided easy-to-read results after only 15 min of incubation.
A limitation of this study is that a relatively small number of isolates were assayed; however, the sensitivity and specificity of the OKNV assay were similar to those reported in previous studies [101112].
Overall, the speed and ease-of-use of the OKNV assay represent significant technical advances; the assay also shows excellent performance for detecting the OXA-48-like, KPC, NDM, and VIM-type carbapenemases from Enterobacteriaceae isolates cultured on both SBA and CHROMagar KPC medium.
Notes
References
1. Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014; 20:821–830. PMID: 24930781.
2. Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013; 4:48. PMID: 23504089.
3. Aguirre-Quiñonero A, Martínez-Martínez L. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother. 2017; 23:1–11. PMID: 27769646.
4. Dortet L, Poirel L, Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother. 2012; 56:6437–6440. PMID: 23070158.
5. Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2015; 70:3014–3022. PMID: 26260131.
6. Glupczynski Y, Evrard S, Ote I, Mertens P, Huang TD, Leclipteux T, et al. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother. 2016; 71:1217–1222. PMID: 26825120.
7. Glupczynski Y, Jousset A, Evrard S, Bonnin RA, Huang TD, Dortet L, et al. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother. 2017; 72:1955–1960. PMID: 28369469.
8. Papadimitriou-Olivgeris M, Vamvakopoulou S, Spyropoulou A, Bartzavali C, Marangos M, Anastassiou ED, et al. Performance of four different agar plate methods for rectal swabs, synergy disk tests and metallo-β-lactamase Etest for clinical isolates in detecting carbapenemase-producing Klebsiella pneumoniae. J Med Microbiol. 2016; 65:954–961. PMID: 27452448.
9. Lee SY, Octavia S, Chew KL. Detection of OXA-carbapenemase-producing Enterobacteriaceae with chromID CARBA SMART screening plate. Pathology. 2019; 51:108–110. PMID: 30454880.
10. Glupczynski Y, Evrard S, Huang TD, Bogaerts P. Evaluation of the RESIST-4 K-SeT assay, a multiplex immunochromatographic assay for the rapid detection of OXA-48-like, KPC, VIM and NDM carbapenemases. J Antimicrob Chemother. 2019; 74:1284–1287. PMID: 30753488.
11. Greissl C, Saleh A, Hamprecht A. Rapid detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in Enterobacterales by a new multiplex immunochromatographic test. Eur J Clin Microbiol Infect Dis. 2019; 38:331–335. PMID: 30448931.
12. Rösner S, Kamalanabhaiah S, Küsters U, Kolbert M, Pfennigwerth N, Mack D. Evaluation of a novel immunochromatographic lateral flow assay for rapid detection of OXA-48, NDM, KPC and VIM carbapenemases in multidrug-resistant Enterobacteriaceae. J Med Microbiol. 2019; 68:379–381. PMID: 30663953.
13. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011; 70:119–123. PMID: 21398074.
14. Jeong SH, Kim HS, Kim JS, Shin DH, Kim HS, Park MJ, et al. Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016; 36:529–535. PMID: 27578505.
15. Shin J, Baek JY, Chung DR, Ko KS. Instability of the IncFII-type plasmid carrying blaNDM-5 in a Klebsiella pneumoniae isolate. J Microbiol Biotechnol. 2017; 27:1711–1715. PMID: 28746989.
16. Korea Centers for Disease Control and Prevention. Korean Antimicrobial Resistance Monitoring System (KARMS) annual report, 2016. Korea Centers for Disease Control and Prevention. 2018.
17. Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2018; 73:909–915. PMID: 29365094.
Fig. 1
Example of results for a KPC-2 and NDM-1 co-producing Citrobacter freundii isolate grown on (I) sheep blood agar and (II) CHROMagar KPC (1, cassette for (OXA)-48-like (O) and KPC (K); 2, cassette for VIM (V) and NDM (N)). In the case of a negative result, only a single line appears at the position of the control band (C).
Abbreviations: OXA, oxacillinase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase.
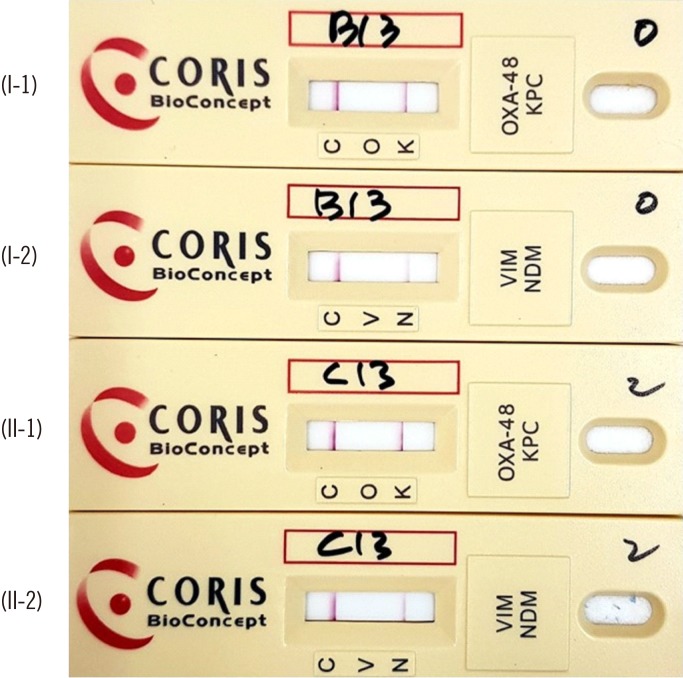
Table 1
Results of the OKNV assay for carbapenem-resistant Enterobacteriaceae isolates
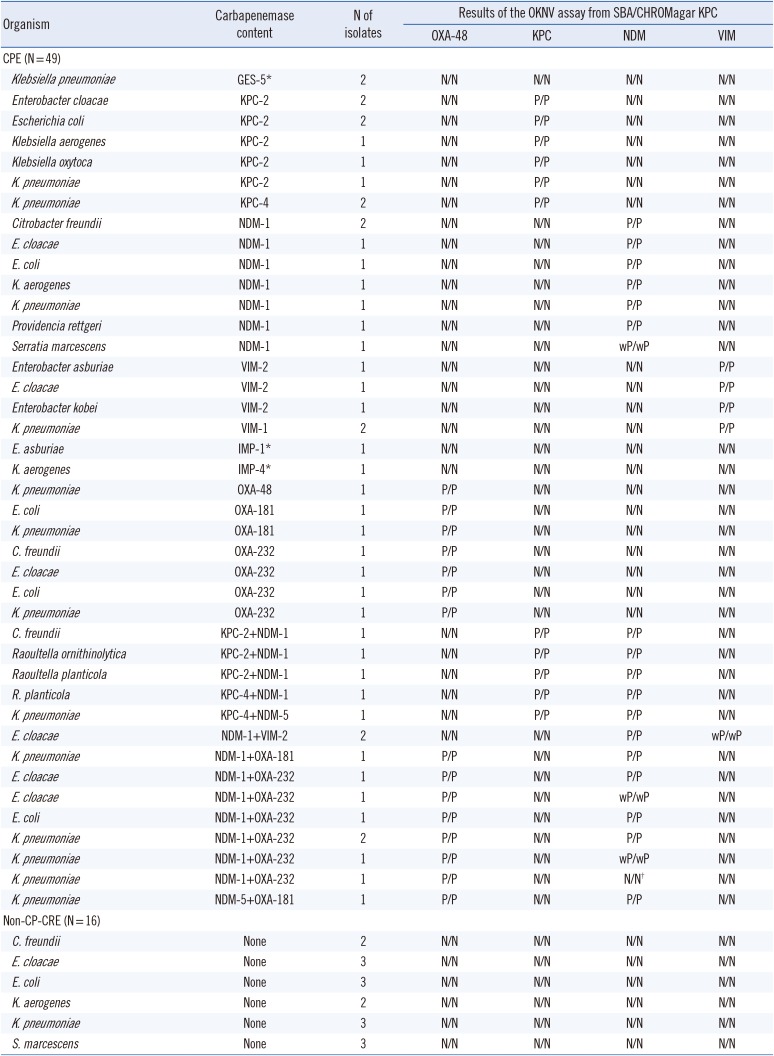
*The OKNV assay can only detect OXA-48, KPC, NDM, and VIM type carbapenemases; †Boldface represents false-negative results.
Abbreviations: OKN, RESIST-4 O.K.N.V; CP-CRE, carbapenemase-producing carbapenem-resistant Enterobacteriaceae; SBA, sheep blood agar; P, positive; wP, weakly positive; N, negative; CPE,carbapenemase-producing Enterobacteriaceae; OXA, oxacillinase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download